Team:Duke/Project/Design
From 2013.igem.org
Experimental Design
- Design enhancer sequences and corresponding artificial transcription factors (ATFs). Check constructs against the yeast genome and transcription factor sequences to ensure independence of the system and limit off-target effects.
- Construct two plasmid libraries: one containing fluorescent reporter genes driven by constitutive promoters, and one containing artificial transcription factor genes (TALEs, dCas9, and sgRNAs) driven by inducible promoters. Enhancer sequences to match each ATF were inserted between the promoter and the gene in the reporter constructs.
- Integrate plasmids into the yeast genome, in pairs of corresponding reporters and ATFs. Use flow cytometry along with control strains to quantifiably test for repressive activity and cooperativity in ATF-enhancer interaction.
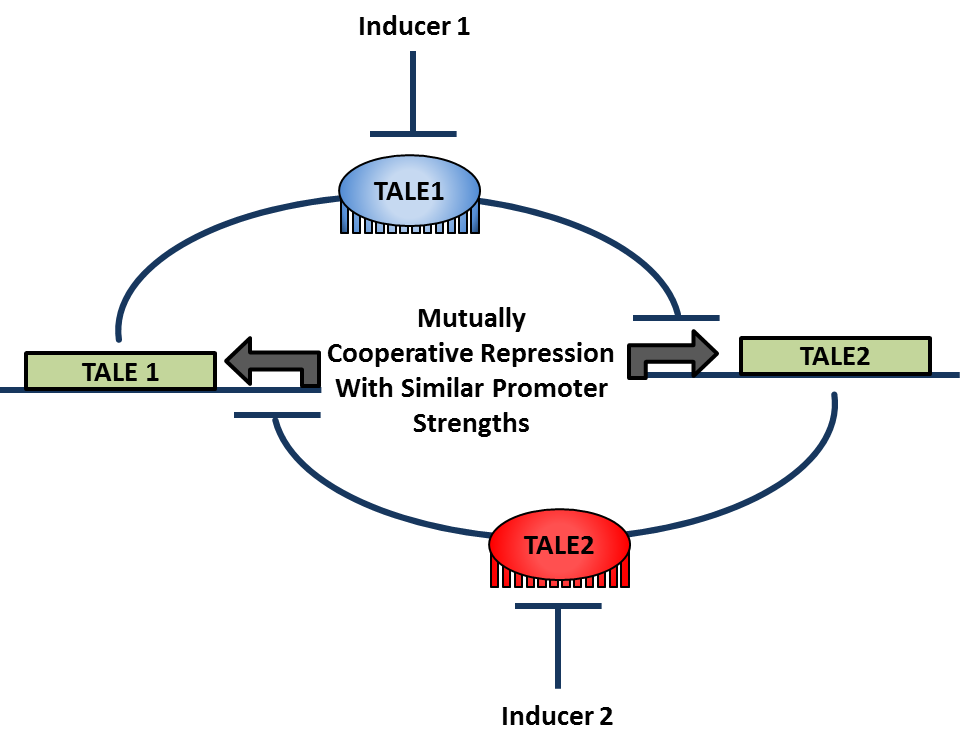
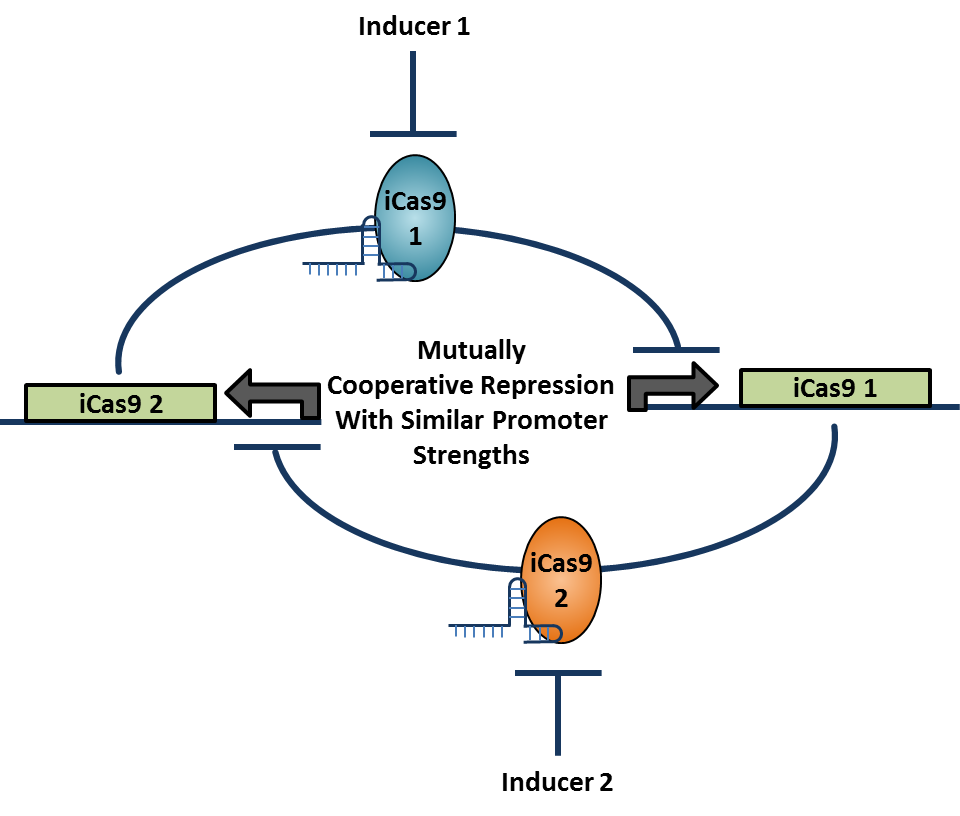
- Select compatible sets of ATF-enhancer pairs, using mathematical modeling results to determine the most robust circuit designs. Design, integrate, and test these circuits for balanced cooperative repression and bistability.
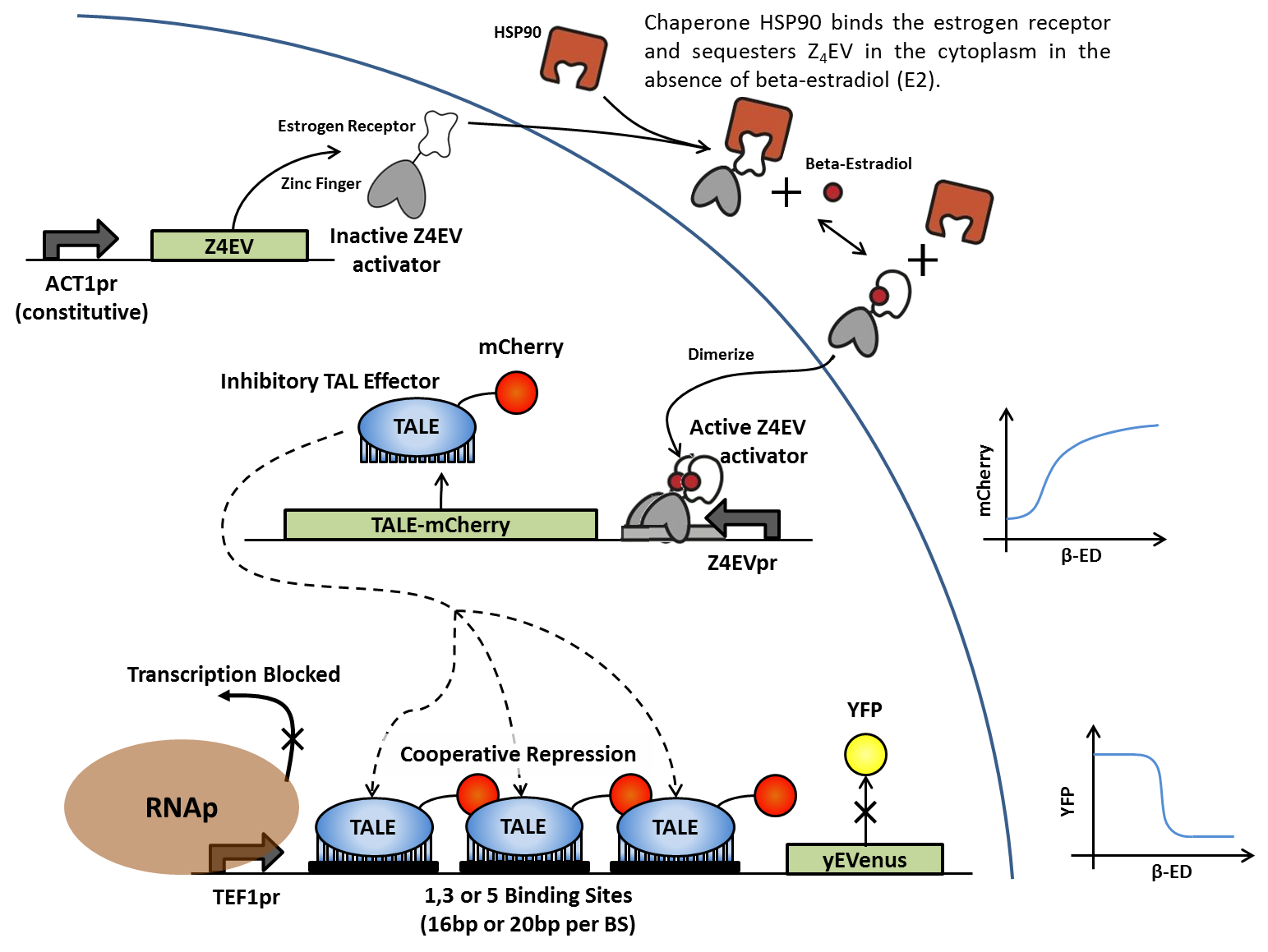
Addition of (β)-estradiol activates transcription of iTAL-mcherry fusion protein, which in turn binds to artificial enhancer sequences in front of the yEVenus reporter gene. Binding of the iTAL to multiple sites between the constitutive promoter and the reporter gene blocks transcription by RNA polymerase and suppresses yEVenus levels in a cooperative fashion. Multiple variations of this construct differ in enhancer length and sequence, as well as number of tandem binding sites. iTALs are designed to match specific enhancer sequences.
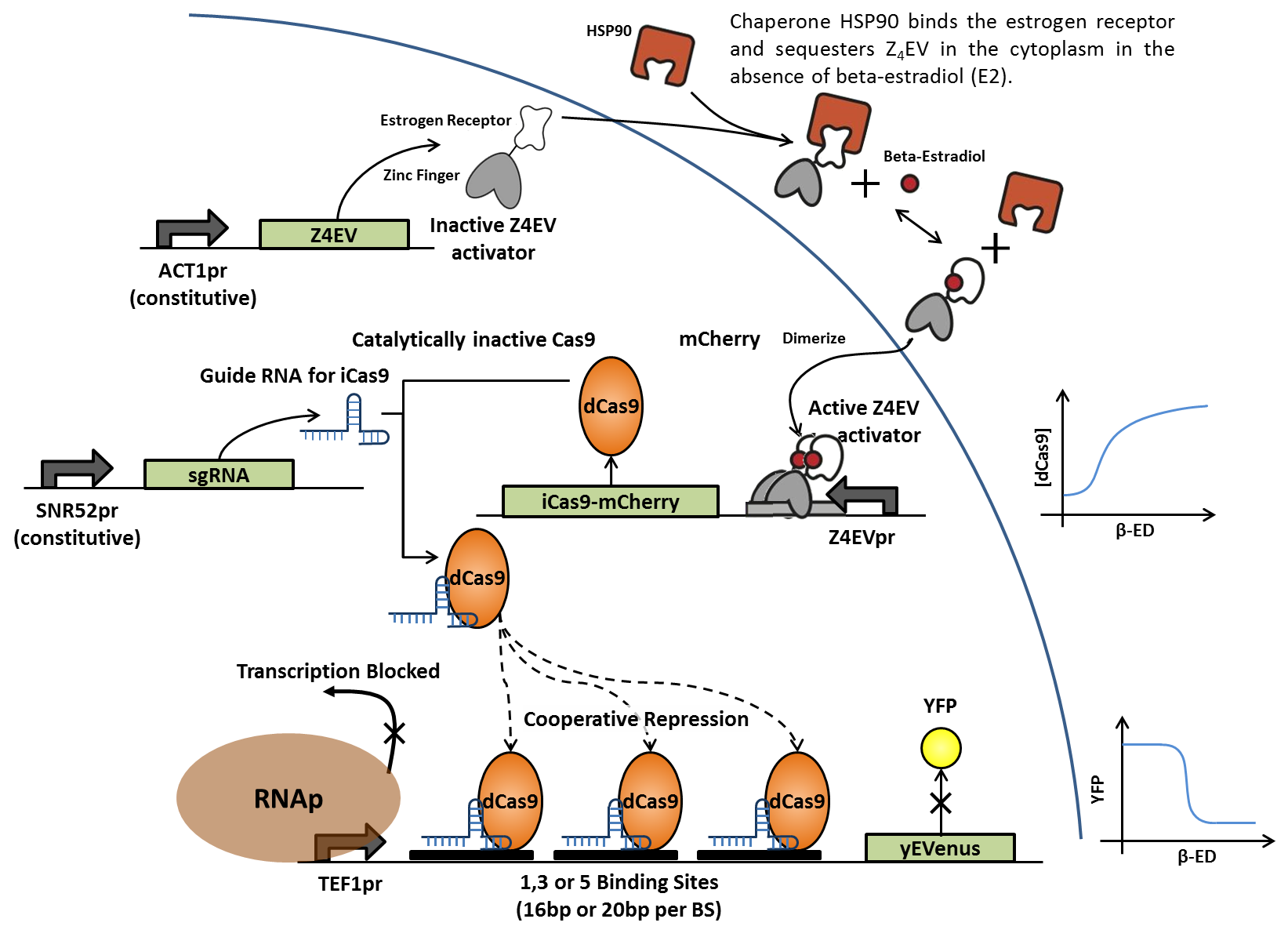
Addition of (β)-estradiol activates transcription of dCas9-mCherry fusion protein, which associates with constitutively expressed, sequence-specific small guide RNAs (sgRNAs). The RNA-protein complex then binds to artificial enhancer sequences in front of the yEVenus reporter gene. Binding of the complex to multiple sites between the constitutive promoter and the repoter gene blocks transcription by RNA polymerase and suppresses yEVenus levels in a cooperative fashion. Multiple variations of this construct differ in enhancer length and sequence, as well as number of tandem binding sites. Each sgRNAs is designed to match a specific enhancer sequence.
References
- McIsaac R.S, Oakes B.L, Wang X, Dummit K.A, Botstein D, Noyes M.B: Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nuc Ac Res 2012, 1-10.
- Murphy K.F, Balazsi G, Collins J.J: Combinatorial promoter design for engineering noisy gene expression. Proc. Nat. Acad. Sci. USA 2007, 104(31):12726–12731.
- Qi L.S, Larson M.H, Gilbert L.A, Doudna J.A, Weissman J.S, Arkin A.P, Lim W.A: Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152:1173-1183.
 "
"