Team:NYMU-Taipei/Modeling/ModSensors
From 2013.igem.org


Contents |
Function of the parts
TrxC promoter is an OxyR-activated promoter. We use TrxC promoter as a sensor/ switch to have the whole circuit opened when Nosema exists, and closed when Nosema is killed.
When Nosema enters the bees, it will trigger bees’ ROS (Reactive Oxygen Species) production, which in turn launches E.coli’s OxyR production. After that ROS and OxyR will form a complex and binds to TrxC promoter. To increase the sensitivity of this switch, we add an additional circuit expressing OxyR regulated by a constitutive promoter to enhance the basal OxyR expression in E.coli.
The purpose of this modeling:
- To know the lag time between sensing the invasion and the production of the killing protein.
- The minimal Oxidative stress that can activate the switch by choosing the proper constitutive promoter for boosting OxyR's concentration
- To know the relation between ROS input and the TrxC promoter open strength.
- To see to which degree will TrxC promoter influence the production of LuxI, LuxR, and TetR, which will influence the second and the third circuit.
It is assumed that the concentration of ROS in bees is relative to the severity of Nosema infection, and that OxyR is so abundant that once ROS appears, it will soon bind to ROS to form OxyR* and reach equilibrium.
Equation1:
PoPSconstitutive = promoter strength of constitutive promoter (J23102)
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know the production rate of mRNAOxyR, and choose the proper constitutive promoter for boosting OxyR's concentration.
Equation2:
RBS = binding site strength
KdegOxyR = degrading constant of OxyR
The aim of the equation is to know the production rate of OxyR and when it can reach the concentration activating TrxC promoter, which can in turn be deduced to get the lag time between sensing the invasion and the production of the killing protein.
Equation3:
KOxyR* = constant rate of ROS+OxyR→OxyR*
The aim of the equation is to know the concentration of OxyR* given the concentration of ROS and OxyR, which will know the relation between ROS input and the TrxC promoter open time.
Equation4:

PoPSconstitutive = promoter strength of constitutive promoter
N = number of plasmid in a single cell
V = volume of a cell
The aim of the equation is to know how TrxC promoter strength (in PoPS) influences the production of TetR, LuxI, and LuxR.
Explanation
In this equation, PoPSconstitutive represents the promoter strength of promoter J23102, which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription.
For the section of the equation, PoPSconstitutive\times\frac{N}{V} represents the synthesizing rate of mRNAOxyR ; -KdegmRNA[mRNAOxyR] represents the degrading rate of mRNAOxyR .
In this equation, RBS represents ribosome binding site strength, which is the affinity of ribosome to the starting site of mRNA.
For the section of the equation, RBS×[mRNAoxyR] represents the synthesizing rate of OxyR; KdegOxyR[xyR] represents the degrading rate of OxyR .
This equation shows the equilibrium of ROS + OxyR → OxyR* (KOxyR* is the rate constant of ROS + OxyR → OxyR*)

In this equation, PoPSTrxC represents the promoter strength of promoter TrxC, which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription;  represents the hill effect of activator OxyR* to TrxC promoter.
represents the hill effect of activator OxyR* to TrxC promoter.
The production rate of mRNATetR, mRNALuxI, mRNALuxR can be written as  , which is under the influence of activator OxyR* binding to TrxC promoter.
, which is under the influence of activator OxyR* binding to TrxC promoter.
Results
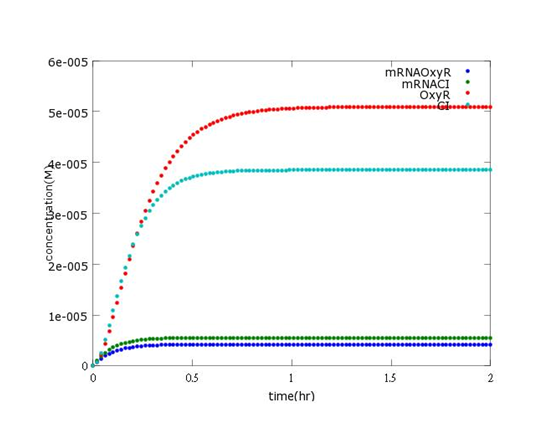
This picture shows the prediction of what concentration of mRNAOxyR, mRNACI, OxyR, CI will reach after every single Bee. coli is born (without Nosema invasion).
As the background of main part wiki has indicated, without Nosema, the second circuit will open and CI will be produced to a certain concentration. This corresponds to the picture above, where CI concentration approaches 3.8×10-5M.
As the assumptions of sensor wiki have written, OxyR is abundant enough to swiftly forms a complex after ROS appears. After that, the complex will reach equilibrium after Nosema infection. As a consequence, we predicted that the concentration will sustain at a constant level, which is 5.1×10-5 in the picture. However, in reality, the final centration of OxyR can reach much more higher than we had expected after a longer period of time. The inaccuracy will be discussed below.
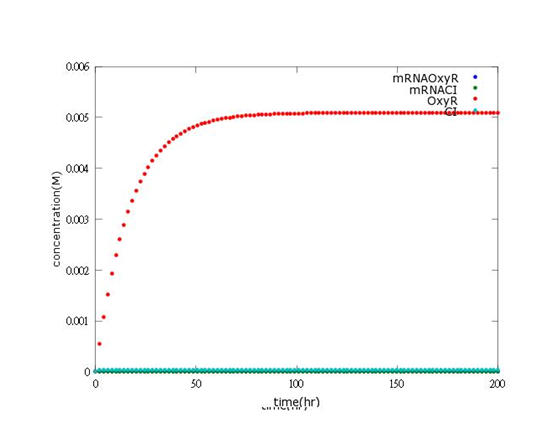
According to the paper data, this picture shows the real concentration of mRNAOxyR, mRNACI, OxyR, CI will reach after every single Bee. coli is born (without Nosema invasion).
The picture indicates that concentration of OxyR is 0.005M, which is much higher than the expectation (5.1×〖10〗^(-5)).
Discussion
The reason why the actual result of OxyR concentration differing from the expected one may be that we did not take the negative feedback of OxyR production.
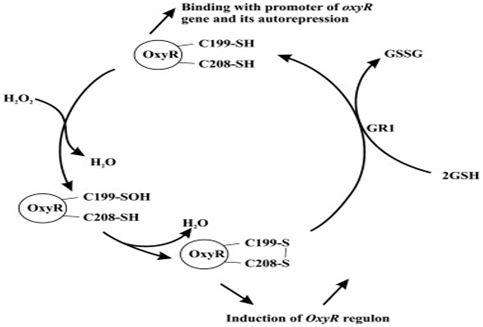
This picture shows OxyR cycle in E. coli:
The top molecule is reduced form of OxyR, whose Cys199 to sulfoxide will be oxidized by hydrogen peroxide (the oxidative reagent as well as a kind of ROS).
After the water exclusion and disulfide bond formation, the reduced form of oxy turns into the oxidized one. Oxidized OxyR is an activator of target promoter, which stimulates downstream gene expression. However, Oxidized form of OxyR protein is transformed into reduced OxyR protein (inactivated) by thioredoxin reductase (GSH). Reduced OxyR protein (the top molecule)provides the autorepression of OxyR gene (a negative feedback).
Parameter
| align="center" !Model | align="center" !Parameter | align="center" !Description | align="center" !Value | align="center" !Unit | align="center" !Reference |
| sensor | KdegOxyR | OxyR degrading rate | 107 | M-1 x min-1 | Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol—disulfide status |
| KdOxyR* | Dissociation constant of OxyR* | 10-7.33 | M | ||
| KOxyR | OxyR producing rate constant | 5.012 x 1014 | X | ||
| nOxyR* | Hill coefficient of OxyR* | Ranging from 0.75~3.5,depends on what kind of ROS it react with | X | OxyR: A Molecular Code for Redox-Related Signaling | |
| N | Copy number | 887(cells containing 25% plasmid bearing cells) | Single piece |
|
 "
"