Team:NYMU-Taipei/Project/Inhibition/Id-Nosema
From 2013.igem.org


Contents |
Identifying Nosema ceranae
Introduction
The identification of N. apis and N. ceranae is important and still needs further research. So far the best differentiation method is the use of species–specific primers; however, the sequence differences are limited to the ribosomal RNA loci, leading to overestimation of Nosema infection rate. Thanks to recent genome study of N. apis and N. ceranae, we proposed novel primers for differentiation of N. spp. based on highly conserved, species–specific protein coding genes rather than species–specific sequences in rRNA genes, which is likely to improve the accuracy of species identification.
Background
N. spp. Differentiation Methods
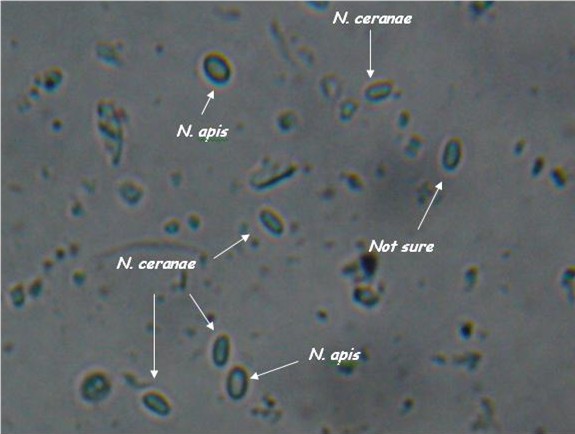
- Light microscopy method: It is based on morphology difference of N. apis and N. ceranae, revealing that spores of N. ceranae are oval or rod shaped like a rice, and also slightly thinner than N. apis. Light microscopy method is convenient but not scientific enough since N. apis bears a striking resemblance to N. ceranae.
- Molecular methods: Species-specific primers are more accurate than light microscopy method. The problem is that so far nearly all protocols based on species–specific sequence differences are limited to the ribosomal RNA loci, which may lead to confusing results due to polymorphisms in and recombination between the multi-copy 16S rRNA genes, and hence overestimate the rate of Nosema infections.
- Our new primers: This year, we created better primers based on recently published, highly conserved and absolutely species–specific sequence differences in protein coding genes. To be more specific, the primers are based on species–specific genes instead of species–specific sequences. As more and more genes of Nosema spp. are revealed, we are able to choose the genes that are not homologs N. apis and N. ceranae for new primers. Our new primers are confirmed by BLAST through nucleotide blast and tblastn using Nucleotide collection (nr/nt) database and Whole-genome shotgun contigis (wgs) database. We propose that our new primers should become the preferred molecular tool for differentiation of Nosema spp. and thus assist in apiculture.
Experiment
Primer Design
1.Common gene of N. spp.--- NcORF_00182 hypothetical protein & NAPIS_ORF01775 chitin synthase d
NcORF_00182 for N. apis
Product=1089bp
NAPIS_ORF01775 N. ceranae
Product=1103bp
2.Specific gene of N. ceranae---NcORF_00714 hypothetical protein
NcORF_00714
Product=861bp for N. ceranae
3.Specific gene of N. apis---NAPIS_ORF02138 class iv chitinase
NAPIS_ORF02138
Product=1320bp for N. apis
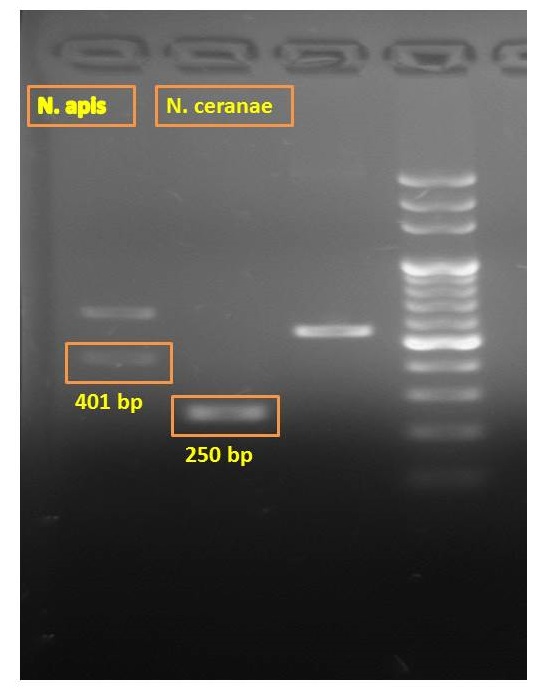
Besides testing our new primers, we also chose traditional primers to double-check whether the bees were infected with N. app. The first step is to do real PCR with primers and spores extracted from infected bees, and then run the electrophoresis to check the length.
Traditional Primer
N. ceranae primers
forward primer:cggataaaagagtccgttacc
reverse primer:tgagcagggttctagggat
Product=250bp for N. ceranae
N. apis primers
forward primer:ccattgccggataagagagt
reverse primer:cacgcattgctgcatcattgac
Product=401bp for N. apis
The electrophoresis result showed that bees were infected with both N. apis and N. ceranae. Another thing worth mentioning is that two bands instead of one band appeared when checking the existence of N. apis. The detection of N. apis is very interesting because many professionals believe that there has no N. apis in Taiwan anymore. In the future, we are planning to do is to compare the outcomes of our new primers with traditional ones to verify the feasibility of the novel method.
Reference
- Gisder, S., & Genersch, E. (May 01, 2013). Molecular differentiation of Nosema apis and Nosema ceranae based on species–specific sequence differences in a protein coding gene. Journal of Invertebrate Pathology, 113, 1, 1-6.[http://www.sciencedirect.com/science/article/pii/S0022201113000165]
- Nabian, S., Ahmadi, K., Nazem, S. M., & Gerami, S. A. (January 01, 2011). First Detection of Nosema ceranae, a Microsporidian Protozoa of European Honeybees (Apis mellifera) In Iran. Iranian Journal of Parasitology, 6, 3, 89-95.[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3279895/]
- Chen, Y., Pettis, J. S., Zhao, Y., Liu, X., Tallon, L. J., Sadzewicz, L. D., Li, R., ... Evans, J. D. (January 01, 2013). Genome sequencing and comparative genomics of honey bee microsporidia, Nosema apis reveal novel insights into host-parasite interactions. Bmc Genomics, 14.[http://www.biomedcentral.com/1471-2164/14/451/]
- Sagastume, S., del, A. C., Martín-Hernández, R., Higes, M., & Henriques-Gil, N. (January 01, 2011). Polymorphism and recombination for rDNA in the putatively asexual microsporidian Nosema ceranae, a pathogen of honeybees. #Environmental Microbiology, 13, 1, 84-95.[http://www.ncbi.nlm.nih.gov/pubmed/21199250]
- Chen, Y. P., Evans, J. D., Murphy, C., Gutell, R., Zuker, M., Gundensen-Rindal, D., & Pettis, J. S. (January 01, 2009). Morphological, molecular, and phylogenetic characterization of Nosema ceranae, a microsporidian parasite isolated from the European honey bee, Apis mellifera. The Journal of Eukaryotic Microbiology, 56, 2.)[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2779023/]
- Huang, W. F., Bocquet, M., Lee, K. C., Sung, I. H., Jiang, J. H., Chen, Y. W., & Wang, C. H. (January 01, 2008). The comparison of rDNA spacer regions of Nosema ceranae isolates from different hosts and locations. Journal of Invertebrate Pathology, 97, 1, 9-13. [http://ansc.niu.edu.tw/files/writing_journal/20/27_2fcb1cab.pdf]
- Bollan, K. A., Hothersall, J. D., Moffat, C., Durkacz, J., Saranzewa, N., Wright, G. A., Raine, N. E., ... Connolly, C. N. (January 01, 2013). The microsporidian parasites Nosema ceranae and Nosema apis are widespread in honeybee (Apis mellifera) colonies across Scotland. Parasitology Research, 112, 2, 751-9. [http://www.ncbi.nlm.nih.gov/pubmed/23180128]
- Huang, W. F., Solter, L. F., Yau, P. M., & Imai, B. S. (January 01, 2013). Nosema ceranae escapes fumagillin control in honey bees. Plos Pathogens, 9, 3.) [http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1003185]
 "
"