Team:Heidelberg/Templates/Materials list
From 2013.igem.org
Kits
| Kit | Supplier | Catalog Number |
|---|---|---|
| MinElute® PCR Purification Kit (250) | QIAGEN | 28006 |
| Plasmid Plus Maxi Kit (25) | QIAGEN | 12963 |
| Plasmid Plus Midi Kit (25) | QIAGEN | 12943 |
| QIAEX II® Gel Extraction Kit (500) | QIAGEN | 20051 |
| QIAGEN® Plasmid Plus Midi Kit (100) | QIAGEN | 12945 |
| QIAquick® Gel Extraction Kit (250) | QIAGEN | 28706 |
| QIAquick® Nucleotid Removal Kit (250) | QIAGEN | 28306 |
| QIAquick® PCR Purification Kit (250) | QIAGEN | 28106 |
| QIAprep® Spin Miniprep Kit (250) | QIAGEN | 27106 |
| QIAprep Spin Miniprep Columns | QIAGEN | 27115 |
Marker
| Marker | Supplier | Catalog Number |
|---|---|---|
| Quick-Load® 2-Log DNA Ladder (0.1-10.0 kb) | New England BioLabs | N3200S |
| Quick-Load® 1 kb DNA Ladder | New England BioLabs | N0468S |
| 50 bp DNA Ladder | New England BioLabs | N3236S |
| Gel loading solution | Sigma-Aldrich Chemie GmbH | G2526-5ML |
Enzymes
| Enzyme | Supplier | Catalog Number |
|---|---|---|
| DreamTaq Green PCR Master Mix (2X) | Thermo Fisher Scientific Biosciences GmbH | K1081 |
| Dream Taq PCR MM | Fermentas Life Sciences | K1071 |
| Gibson Assembly® Master Mix | New England Biolabs | E2611 S |
| Lysozyme from chicken egg white | Sigma-Aldrich Chemie GmbH | L4919-500MG |
| Phusion Flash High-Fidelity PCR MAster Mix | Biozym Scientific GmbH | F-548L |
| Phusion® High-Fidelity PCR Master mix | New England Biolabs | M0531 L |
| T4 DNA Ligase | New England Biolabs GmbH | M0202 S |
| 2x PCR Master mix Solution (iTaq) | HISS DIAGNOSTICS GmbH | 25028 |
| Pronase from Streptomyces griseus | Sigma-Aldrich Chemie GmbH | P6911-100MG |
Restriciton Enzymes
| Enzyme | Supplier | Catalog Number |
|---|---|---|
| BamI | New England Biolabs | R3136 S |
| BgIII | New England Biolabs | R0144 S |
| DpnI | New England Biolabs | R0176 S |
| EcoRI | New England BioLabs | R0101S |
| EcoRI-HF | New England Biolabs | R3101 |
| HindIII-HF | New England Biolabs | R3104 S |
| KpnI-HF | New England Biolabs | R3142 S |
| MfeI-HF | New England Biolabs | R3589 S |
| NheI-HF | New England BioLabs | R3131 S |
| NotI-HF | New England BioLabs | R3189 S |
| PacI | New England Biolabs | R0547 S |
| PstI-HF | New England Biolabs | R3140 S |
| PvuI-HF | New England BioLabs | R3150S |
| SalI-HF | New England Biolabs | R3138 S |
| SpeI-HF | New England BioLabs | R3133 S |
| XbaI | New England BioLabs | R0145 S |
Bacterial Strains
| Strain | Supplier | Catalog Number |
|---|---|---|
| Delftia acidovorans DSM-39 | DSMZ | DSM 50251 |
| Delftia acidovorans SPH-1 | DSMZ | DSM 14801 |
| E. coli DH10ß | New England Biolabs | C3019 |
| E. coli Top10 | invitrogen | C404010 |
| Photorhabdus laumondii luminescens | DSMZ | DSM 15139 |
| Streptomyces lavendulae lavendulae | DSMZ | DSM 40708 |
| Streptomyces mobaraensis | DSMZ | DSM 40903 |
Antibiotics and Media Supplements
| Antibiotic | Supplier | Catalog Number | Concentration stock solution | Dilution | Solvent |
|---|---|---|---|---|---|
| Ampicillin anhydrous crystalline | Sigma-Aldrich Chemie GmbH | A9393-5G | 100mg/ml | 1:1000 | H2O |
| Ampicillin sodium crystalline | Sigma-Aldrich Chemie GmbH | A9518-5G | 100mg/ml | 1:1000 | H2O |
| Chloramphenicol crystalline | Sigma-Aldrich Chemie GmbH | C0378-5G | 30mg/ml | 1:3000 | Ethanol |
| Kanamycinsulfat Gemisch der Komponentena | Sigma-Aldrich Chemie GmbH | 60615-5G | 50mg/ml | 1:1000 | H2O |
| Tetracycline | Sigma-Aldrich Chemie GmbH | T7660 | 10mg/ml | 1:1000 | Ethanol |
| Propionic acid sodium insect cell*culture | Sigma-Aldrich Chemie GmbH | P5436-100G | 100mM | 10mM | H2O |
| Bacitracin | Sigma-Aldrich Chemie GmbH | B0125-50KU | - | - |
Medium
| Media | Supplier | Catalog Number |
|---|---|---|
| SOC Outgrowth Medium | New England Biolabs GmbH | B9020 S |
| LB BROTH BASE | Th. Geyer GmbH & Co KG | SA/L3022/001000 |
| LB Broth powder | Sigma-Aldrich Chemie GmbH | L3022-1KG |
| M9 minimal salts | SERVA | 48505.01 |
Electrophoresis
| Agarose molecular Biology Reagent | Th. Geyer GmbH & Co KG | SA/A9539/000050 | 0,5% | H2O |
| Agarose for routine use | Sigma-Aldrich Chemie GmbH | A9539-100G | - | - |
Miscellaneous Primers
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| VF2 | - | for screening in standard BB backbone- binds on the Backbone before Insert | TGCCACCTGACGTCTAAGAA |
| VR | - | for screening in standard BB backbone- binds on Backbone behind Insert | ATTACCGCCTTTGAGTGAGC |
Primers and Oligos
Delftibactin
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| DN01:delH_f1_PacI_fw | - | fw Primer for DelH-fragment1 with RBS and PacI-restriction site | TTTT TTAATTAA TCACACAGGAAAGTACTAG ATGGACCGTGGCCGCCTGC GCCAAATCG |
| DN02:delH_f1_SalI_rev | - | - | TTTT GTCGACCAACACCTGTGCCTGC |
| DN03:delH_f2_SalI_fw | - | - | TTTT GTCGACTGGATGGAGCCTGGTGAAAG |
| DN04:delH_f2_KpnI_rev | - | - | TTTT GGTACC TCAGTCCAGCGCGTACTCCAG |
| DN05:AraCbb_KpnI_fw | - | amplifying the Backbone for DelH (pSB6A1-AraC-lacZ) | TTTT GGTACC AAAGAGGAGAAATACTAGATGACCATG |
| DN06:AraCbb_PacI_rev2 | 15-05-2013 | amplifying the Backbone for DelH (pSB6A1-AraC-lacZ) | TTTT TTAATTAA GCTAGCCCAAAAAAACGGTATG |
| DN07:Screen_delH_rev | 15-05-2013 | for screening if DelH is present - binds on the very beginning of DelH | CTTTCCTCGAACACCGTGCGCAG |
| DN08:DelH_EcoRI_rev | - | rev_Primer for DelH Fragment f1a | CTCGTCGCCATGGACCAGGCAG |
| DN09:DelH_f1_fw_long | 2013-06-11 | for amplifying DelH-1a from the genome: doesn't work | ATGGACCGTGGCCGCCTGCGCCAAATCG |
| DN10:DelH_f1_fw_short | 2013-06-11 | for amplifying DelH-1a from the genome: doesn't work | ATGGACCGTGGCCGCCTGC |
| DN11:DelH_f1_fw_short2 | 2013-06-11 | for amplifying DelH-1a from the genome: works!!!! | GCCGCCTGCGCCAAATCG |
| DN12:DelH_f1_PacI_fw_short | 2013-06-11 | for amplifying DelH-1a from the genome: doesn't work | TTTTTTAATTAATCACACAGGAAAGTAC TAGATGGACCGTGGCCGCCTGC |
| DN13:Screen_DelH_fw | 15-05-2013 | PCR Screening for presence of DelH insert | GTAAACCCACTGGTGATACCATTC |
| FS_01: pSB4K5_DelA_rv | 20-13-06-28 | Amplification of pSB4K5 from the iGEM Distribution Gibson Primer with overhang to DelA introducing the RBS BBa_B0035 | TCGCGGCGATCCGGTACTGCGCCTCTGTT GAACATCTGATATTCTCCTCTTTAATCG ACAGATTGTGTGAAATTGTTATCCGCTCAC |
| FS_02: DelAG_1_fw | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | TTCAACAGAGGCGCAGTACCGGATC |
| FS_03: DelAG_1_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | GTCGGAGACGATGTGGTGCATCAC |
| FS_04: DelAG_2_fw | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | CTGCAGGCCAATGAGCACATCCTG |
| FS_05: DelAG_2_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | CACAGGTGGTAGATGGCGTC |
| FS_06: DelAG_3_fwG | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | ATTGCGAGGACTTGCTCGATG |
| FS_07: DelAG_3_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | TTTGCTGCAGCGCCAGCACATCGAG |
| FS_08: DelAG_4_fw | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | GTACGGCCTATCACATCAGCG |
| FS_09: DelAG_4_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | GAAGCTCAGCAGGTTGGGCGAGACG |
| FS_10: DelAG_5_fw | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | GAATTTTGTTCCACCACCTGCTG |
| FS_11: DelAG_5_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer with overhang to DelOP | CTTGAGCAGGCGCAGTACCTCGGAGGG CGGTCGGCTGGCGTTTTCCATGATT CAGGTTTCCTGTGTGAAGCTCATCTCAGATA TCTCCCAGAGTTTCGAGAAAG |
| FS_11: DelAG_5_short_rv | 2013-05-07 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | TCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_12: DelOP_fw | 2013-06-28 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | GAATCATGGAAAACGCCAGCCGAC |
| FS_13: DelOP_rv | 2013-06-28 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer with overhang to DelL | CAATGTTGGAGGGGCCGAAGCCGATGCCGATC AGCGGGTGGGTTTGCATGGAAGGTC CTTTCATTGGGTCGATGCGTCCAGTGT CACACCGTGGTGTCTGCAGGCG |
| FS_13: DelOP_short_rv | 2013-05-07 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer with overhang to DelL | TCACACCGTGGTGTCTGCAGGCG |
| FS_14: DelL_fw | 2013-06-28 | Amplification of DelL from Delftia acidovorans genome Gibson Primer | CAAACCCACCCGCTGATCGGCATC |
| FS_15: DelL_mRFP_pSB4K5_rv | 2013-06-28 | Amplification of DelL Gibson Primer with overhang to BBa_J04450 | GAAACGCATGAACTCTTTGATAACGTCT TCGGAGGAAGCCATCTAGTATTTCTCCT CTTTCTCTAGTATCAGTCCTGCAGCG CCAGCTGTTCTGTG |
| FS_15: DelL_mRFP_pSB4K5__short_rv | 2013-05-07 | Amplification of DelL Gibson Primer with overhang to BBa_J04450 | TCAGTCCTGCAGCGCCAGCTGTTCTGTG |
| FS_16: mRFP_pSB4K5_fw (Del) | 2013-06-28 | Amplification of pSB4K5 from iGEM Distribution Gibson Primer | GCTTCCTCCGAAGACGTTATC |
| FS_20: DelF_fw | 2013-07-13 | Amplification of DelF from Delftia acidovorans genome Gibson Primer | GACTTGCTCGATGCGGTGCAG |
| FS_21: DelF_fw | 2013-07-13 | Amplification of DelF from Delftia acidovorans genome Gibson Primer | GACGCCATCTACCACCTGTG |
| FS_22: DelOP_short_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome inlcuding the recently predicted endogenous Promotor for DelOP Gibson Primer | GATGACGCAGGGCGGCGGAATTTGTTCATC |
| FS_23: DelG_long_rv | 2013-07-13 | Amplification of DelG from Delftia acidovorans genome Gibson primer with overhang to DelOP element including the recently predicted endogenous Promotor | GATGAACAAATTCCGCCGCCCTGCGTCA TCTCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_24: DelAE_rv | 2013-07-13 | Amplification of DelAE from Delftia acidovorans genome Gibson Primer | CAGAAGAATTCCCAGAAGGAGATGTCGAAG |
| FS_25: DelEF_fw | 2013-07-13 | Amplification of DelEF from Delftia acidovorans genome Gibson Primer | ACACGGTGCTGCAGAAAACGCCCTTC |
| FS_26: DelFG_rv | 2013-07-13 | Amplification of DelFG from Delftia acidovorans genome Gibson Primer | GAATTCATCCACGATGATCTGCATG |
| FS_27: DelOP_rv | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | CTTTGGGCCGTGCCGGTTTTTGAGATAC |
| FS_28: DelOP_rv | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | GTTTTTGAGATACGCGCGTTGTCAC |
| FS_29: DelOP_rv | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | TTCCCCCTCTCTTTCTCGCTTC |
| FS_30: DelOP_rv | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | CCGCTTCCCCCTCTCTTTCTCGCTTC |
| FS_31: DelOP_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | GTTGGCGAGTTCAAGAAATG |
| FS_32: DelOP_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | TCCTTCAGGTGTGCGGCAGACAAG |
| FS_33: DelOP_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | TTCTTCGTGATGACGCAGGGCGGCGGAATTTGTTC |
| FS_35: DelG_fw | 2013-08-05 | Amplification of DelG from Delftia acidovorans genome Gibson Primer | CATGCAGATCATCGTGGATGAATTC |
| FS_45: pSB4K5_fw | 2013-08-02 | Amplification of pSB4K5 from iGEM Distribution Gibson Primer without mRFP | CCAGGCATCAAATAAAACGAAAG |
| FS_46: DelL_rv | 2013-08-02 | Amplification of DelL from Delftia acidovorans genome Gibson Primer creating overlap to pSB4K5 without mRFP | TCAGTCCTGCAGCGCCAGCTGTTCTG TGCTTTCGTTTTATTTGATGCCT |
| FS_47_screening_BB_AF_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTGGCCGATTCATTAATGC |
| FS_48_screening_BB_AF_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TAACGGTATCGGTATCGCTTTG |
| FS_49_screening_AFI_AFII_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTTCTCTGGAAGATGGATAC |
| FS_50_screening_AFI_AFII_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTGACGAAAAAGCCGACCAC |
| FS_51_screening_AF_FG(21-26)_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TGGATATCGACTGGACTGCCTG |
| FS_52_screening_AF_FG(21-26)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TGCACCACATCGACGAAACGG |
| FS_53_screening_FG(21-26)_G_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTACGGCCTATCACATCAGCG |
| FS_54_screening_FG(21-26)_G_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GAACCTGGGTGTTCACGAAAAAGCC |
| FS_55_screening_G_OP_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTATCTCTACATGCATCGCTAC |
| FS_56_screening_G_OP_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | AGGACATTTTCCGCACCCCG |
| FS_57_screening_G_OP_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GCTGGCGTTTTCCATAAG |
| FS_58_screening_OP_L_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GAACAACTTCCAGCACAGCCTGTTC |
| FS_59_screening_OP_L_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CGTTGAAGATTTCGTTGACG |
| FS_60_screening_L_BB_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CATCTTCAAGGTGTTCTATGAAC |
| FS_61_screening_L_BB(with_mRFP)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CAGTTTAACTTTGTAGATGAAC |
| FS_66: DelH_rv | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | TGGGCATTCACCGCATCGATC |
| FS_67: DelH_fw | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | CTTCACGTTGATTGCGCATG |
| FS_68: DelH_rv | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | CAGAAGAACTCCCAGACCGAC |
| FS_69: DelH_fw | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | GACACCGTTCAGCTTCGATG |
| FS_70: DelH_rv | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | GAAGCTGCTCCGCTGATAGAT |
| FS_71: DelH_fw | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | ATGTGCTGTCGCTCAAGATG |
| FS_72_SR_02_fw | 2013-08-30 | Screening of pFHFSN | ATGTGCTGTCGCTCAAGATG |
| FS_73_SR_03_fw | 2013-08-30 | Screening of pFHFSN | GTGCTGTTTGGCCGTATG |
| FS_74_SR_04_fw | 2013-08-30 | Screening of pFHFSN | ATCAGGTGCTGAGCTACGAC |
| FS_75_SR_05_fw | 2013-08-30 | Screening of pFHFSN | CTGTTCATCAACACCTTGCC |
| FS_76_SR_06_rv | 2013-08-30 | Screening of pFHFSN | GAAGACAGTCATAAGTGCGGC |
| FS_77_rv | 2013-09-11 | Gibson-Primer rev, Amplficiation of the Backbone pSB6A1 with overlap to the RBS BBa_B0034 and the lacI-promotor, it creates an overlap to the beginning of DelH | GCGATTTGGCGCAGGCGGCCACGGTC CATCTAGTATTTCTCCTCTTTC |
| FS_78_rv | 2013-09-26 | Gibson-Primer rev, Amplficiation of the Backbone pSB6A1 introducing the RBS BBa_B0032 and the promotor BBa_J23114 and creating an overlap to the first fragment of DelH amplified with primer DN_11 | GATTTGGCGCAGGCGGCCACGGTCCA TCTAGTACTTTCCTGTGTGACTCTAG AGCTAGCATTGTACCTAGGACTGAGCTAG CCATAAACTCTAGAAGCGGCCGCGAATTC |
| FS_84_fw | 2013-09-26 | Gibson-Primer fw, Amplficiation of the first fragment of DelH introducing the RBS BBa_B0032 and creating an overlap to primer FS_85 thereby partially introducing the promotor BBa_J23114 | GCTCAGTCCTAGGTACAATGCTAGCT CTAGAGTCACACAGGAAAGTACTAGA TGGACCGTGGCCGCCTGCG |
| FS_85_rv | 2013-09-26 | Gibson-Primer rev, Amplficiation of the Backbone pSB6A1, partially introducing the promotor BBa_B0032 with overlap to primer FS_84 and therefore the promotor BBa_J23114, it creates an overlap to the beginning of DelH | GCGATTTGGCGCAGGCGGCCACGGTCC ATCTAGTATTTCTCCTCTTTC |
| FS_86_rv | 2013-09-27 | Gibson-Primer rev, Amplficiation of the Backbone pSB4K5 without any promotor introducing a KpnI cutting site for restriction cloning, creates an overlap to DelH and will be used for the ccdB strategy | GGCGATTTGGCGCAGGCGGCCACGG TCCATGTACTTCGAGTCACTAAGGGCTAAC |
| FS_87_fw | 2013-09-27 | Gibson-Primer fw, Amplficiation of the Backbone pSB6A1 introducing a BamHI cutting site for restriction cloning and creating an overlap to the last fragment of DelH | CGCTGGAGTACGCGCTGGACTGA GATCCCAGGCATCAAATAAAACG |
| FS_90_fw | 2013-09-27 | Gibson-Primer fw, Amplficiation of the ccdB cassette from the template pDonorPlasmid introducing a KpnI cutting site for restriction cloning, creates an overlap to the promotor BBa_J23114 and will be used for the ccdB strategy | CTCAGTCCTAGGTACAATGCTAGCTCTAGA GTCACACAGGAAAGCAGTACACTGGCT GTGTATAAGGGAG |
| FS_93_rv | 2013-09-27 | Gibson-Primer rev, Amplficiation of the ccdB cassette from the template pDonorPlasmid introducing a BamHI cutting site for restriction cloning, creates an overlap to the backbone pSB6A1 and will be used for the ccdB strategy | GTTCACCGACAAACAACAGATGA TCCGCGTGGATCCGGCTTAC |
| FS_94_fw | 2013-09-27 | Primer fw, Amplficiation of the backbone pSB6A1, will be used for the ccdB strategy | ATCTGTTGTTTGTCGGTGAACGC |
| HM01:DelH_EcoRI_fw | - | fw_Primer for DelH Fragment f1b | GCATTGGAGCCTCAATGGCAAGTC |
| HM02:DelH_Gib1.1_rev | 2013-07-09 | Gibson-Primer DelH | TGCTGCGCCTGCATACGGCCAAACA |
| HM03:DelH_Gib1.2_fw | 2013-07-09 | Gibson-Primer DelH | AGCGGCAGGGACGACGTGGT |
| HM04:DelH_Gib1.2_rev | 2013-07-09 | Gibson-Primer DelH | CATAGAGGTTGTAGAGA |
| HM05:DelH_Gib2.1_fw | 2013-07-09 | Gibson-Primer DelH | AGAACGCCGTCTTCAGGCTCCTG |
| HM06:DelH_Gib2.1_rev | 2013-07-09 | Gibson-Primer DelH | CAATGCTTTG CCGCTCGAA |
| HM07:DelH_Gib2.2_fw | 2013-07-09 | Gibson-Primer DelH | TCGCCACGGCAGCTGTTCGA |
| HM08:DelH_Gib2_end_rev | 2013-07-09 | Gibson-Primer DelH | TCAGTCCAGCGCGTACTCCAG |
| HM09:AraC_RBS_Delh_rev | 2013-07-09 | Gibson-Primer rev, introduces a new RBS and has the AraC-promotor and the beginning of DelH | TTGCAAAGCGCTCGGCGATTTGGCGCAGGCG GCCACGGTCCATTTAACTTTCTCCTC TTTAATACTTTGAGCTAGCCCAA AAAAACGGTATGGAGAAACAGTAGAGAGTT |
| HM10:RBS_lacZ | 2013-07-09 | Gibson-Primer fw for the pSB6A1 Backbone with the end of DelH, RBS(1) and the beginning of lacZ | TGGAGTACGCGCTGGACTGA TCTAGAG AAAGAGGAGAAA TACTAG ATGACCATGATTA |
| HM11:lacI_RBS(1)_DelH_rev | 2013-07-24 | Gibson-Primer rev, amplify the Backbone with overlap with the RBS and the lacI-promotor and it creates and overlap to the start of DelH | TCGGCGATTTGGCGCAGGCGGCCACGGTCC ATCTAGTATTTCTCCTCTTTCTCTAGTATGTGTG |
| HM12:DelH_RBS(1.2)_mRFP_fw | 2013-07-24 | Gibson-Primer fw for the pSB6A1 Backbone with the end of DelH, introducing a new RBS(new) and the beginning of mRFP | ATTGGCGCTGGAGTACGCGCTGGACTG ATCAAAGTATTAAAGAGGAGAAAGT TAAATGGCTTCCTCCGAAGACGTTATCAAAGAG |
| HM13:Screen_DelH_end_fw | 2013-08-16 | New screening primer for the end of DelH together with the VR2 primer from the registry | TTTCTGACGACCCTGCACCTGAAG |
| HM14:DelH_tetR_fw | 2013-08-16 | Gibson-Primer DelH-tetR: amplifies the tetracycline resistance from the pSB1T3 Backbone and creates an overlap to the end of DelH | ATTGGCGCTGGAGTACGCGCTGGACTGA ATGAAGTTTTAAATCAATCTAAAG |
| HM15:tetR_stop_BB_rev | 2013-08-16 | Gibson-Primer tetR-pSB6A1: amplifies the tetracycline resistance and creates an overlap with the Terminator of the Backbone pSB6A1 | CGACTGAGCCTTTCGTTTTATTTGATGCCTGGC CTCGTGATACGCCTATTTTTATAGG |
| HM16:tetR_pSB6A1_fw | 2013-08-16 | Gibson-Primer DelH, amplifies the Backbone pSB6A1 creating an overlap with the tetracycline resistance | AAAAATAGGCGTATCACGAG GCCAGGCATCAAATAAAACGAAAGGCTCAG |
| HM17:DelH_Terminator_BB_fw | 2013-08-16 | Gibson-Primer fw for the pSB6A1 Backbone (binding the terminator) and creating an overlap with the end of DelH | ATTGGCGCTGGAGTACGCGCTGGACTGA AGGCATCAAATAAAACGAAAGGCTCAG |
| HM20:BB_HPLC_rev | 11-09-2013 | HPLC version of HM11 Gibson-Primer rev, amplify the Backbone with overlap with the RBS and the lacI-promotor and it creates and overlap to the start of DelH | GATTTGGCGCAGGCGGCCAC GGTCCATCTAGTATTTCTCCTCTTTC |
| HM21:fw_lacI_BbsI_Xba | 2013-09-15 | Forward primer for cutting out mutated fragment for mutagenesis | TTTTGAAGACAA CTAGGCAATACGCAA |
| HM22:rev_RBS | 2013-09-15 | Reverse Primer in RBS for mutagenesis | TTTTGAAGACAA CTCTTTCTCTAGTATGTGTGAAATTG |
| HM23:fw_RBS | 2013-09-15 | Forward Primer in RBS for mutagenesis | TTTTGAAGACAA AGAGGAGAAATACTAGATGGACCGTGGC |
| HM24:rev_BbsI_MfeI | 2013-09-15 | Reverse primer for cutting out mutated fragment for mutagenesis | TTTTGAAGACAA AATTGGACAGCGCGGCATGCCGGTTG |
| IK01:pLF03_integr_argK_fw | 2013-06-12 | For verification of correct genomic integration of pLF03 into BAP1 via colony-PCR. Primer against BAP1 genome. | GCTGATGGAAGTGGCTGATCTGATC |
| IK02:pLF03_integr_pET21c_rev | 2013-06-12 | For verification of correct genomic integration of pLF03 into BAP1 via colony-PCR. Primer against pLF03 backbone (pET-21c). | TCCGCTCACAATTCCCCTATAGTG |
| IK03:pLF03_integr_argK_rev | 2013-06-18 | For verification of correct genomic integration of pLF03 into BAP1 via colony-PCR. Primer against BAP1 genome for positive control (to be used with IK01 or IK04). | GATAAATTCACTGAGCTGCCGCAG |
| IK04:pLF03_integr_argK_fw | 2013-06-18 | For verification of correct genomic integration of pLF03 into BAP1 via colony-PCR. Primer against BAP1 genome. Alternative forward primer in case IK01 does not work. | GCGGGAATTAATGCTGTTATGCGAAG |
| IK05:pLF03_integr_ygfG_fw | 2013-06-18 | For verification of correct genomic integration of pLF03 into BAP1 via colony-PCR. Primer against BAP1 genome, namely ygfG, which should be replaced by the construct. | GCGGTCATTGAGTTTAACTATGGCC |
| IK06:pLF03_integr_ygfG_rev | 2013-06-18 | For verification of correct genomic integration of pLF03 into BAP1 via colony-PCR. Primer against BAP1 genome, namely ygfG, which should be replaced by the construct. | AACATGGTTGAGGATGCCGACAGC |
| IK07:ygfG21C1 | 2013-06-25 | For insertion of methylmalonyl-CoA synthesis pathway into E. coli. Extended ygfG21C1 with higher melting temperature. | TCACCGCGCACCGGCCTGCGGCAGCTCAGTGAATTTATCC AGATCTCGATCCCGCGAAATTAATAC |
| IK08:ygfG21C2 | 2013-06-25 | For insertion of methylmalonyl-CoA synthesis pathway into E. coli. Extended ygfG21C2 with higher melting temperature. | GTTATGCTGGATAATTTCTGCCGCTTCATTGGCGGTCATC CAAAAAACCCCTCAAGACCCGTTTAG |
| IK24:pccB_fw | 2013-07-16 | Colony-PCR of methylmalonyl-CoA synthesis pathway | CGTATCGAGGAAGCGACGCAC |
| IK25:pccB_rev | 2013-07-16 | Colony-PCR of methylmalonyl-CoA synthesis pathway | GGTGATGAACATGTGGCTGGTCTG |
| IK26:BBa_I746200_fw | 2013-07-19 | Forward primer for permeability device | ATGAACAAGAAGATTCATTCCCTGGCCTTG |
| IK27:BBa_I746200-BBa_B0029_rev | 2013-07-19 | Reverse primer for permeability device with partial Gibson overhang for BBa_B0029 RBS | GAATACCAGT TCAGAAGTGGGTGTTTACGCTCATATAC |
| IK28:pccB-BBa_B0029-BBa_I746200_fw | 2013-07-19 | Forward primer for pccB with BBa_B0029 RBS and Gibson overhang for permeability device | ATATGAGCGTAAACACCCACTTCTGAACTGGTATTCACACAGGAAACCTACTAG ATGTCCGAGCCGGAAGAGC |
| IK29:accA2-BBa_B0030_rev | 2013-07-19 | Reverse primer for accA2 with partial Gibson overhang for BBa_B0030 RBS | TAATGAAGTTG CTAGTGATTCTCGCAGATGGC |
| IK30:sfp-BBa_B0030-accA2_fw | 2013-07-19 | Forward primer for sfp with BBa_B0030 RBS and Gibson overhang for accA2 | TCCGGCGCCGCCATCTGCGAGAAT CACTAGCAACTTCATTAAAGAGGAGAAATACTAG ATGAAGATTTACGGAATTTATATGGAC |
| IK31:sfp_rev | 2013-07-19 | Reverse primer for sfp | |
| IK32:BBa_J04450-sfp_fw | 2013-07-19 | Forward primer for mRFP-containing backbones with Gibson overhang for sfp | ATCACAATGGTCTCGTACGAAGAGCTTTTATAA TACTAGAGCCAGGCATCAAATAAAACG |
| IK33:BBa_J04450-BBa_I746200_rev | 2013-07-19 | Reverse primer for mRFP-containing backbones with Gibson overhang for permeability device | ACAAGGCCAGGGAATGAATCTTCTTGTTCAT CTAGTATTTCTCCTCTTTCTCTAGTATG |
| IK34:pccB_fw | 2013-07-19 | Forward primer for pccB | ATGTCCGAGCCGGAAGAGC |
| IK35:BBa_J04450-pccB_rev | 2013-07-19 | Reverse primer for mRFP-containing backbones with Gibson overhang for pccB | ATGTCGGGCTGCTGCTCTTCCGGCTCGGACAT CTAGTATTTCTCCTCTTTCTCTAG |
| IK36:pLF03_seq_rev | 2013-07-26 | reverse primer for pLF03 for sequencing | CCGGTATCAACAGGGACACCAG |
| IK37:pLF03-catR_seq_rev | 2013-07-26 | reverse primer for pLF03 sequencing | CATTGAGCAACTGACTGAAATGCCTC |
| IK38:pIK1_fw | 2013-08-30 | Forward mutagenic primer for pIK1 | ATTCCCTGGCCTTGT T GGTCAATCTGGGGATTTATG |
| IK39:pIK_rev | 2013-08-30 | Reverse mutagenic primer for pIK1 | CCCAGATTGACC A ACAAGGCCAGGGAATGAATCTTC |
| IK40:pIK2-BBa_J23114-BBa_B0030_rev | 2013-09-06 | reverse primer for pIK2 with Gibson overhang for BBa_J23114-BBa_B0030 | TTTAATCTCTAGAGCTAGCATTGTACCTAGGACTGAGCTAGCCATAAA CTCTAGTAGAGAGCGTTCAC |
| IK41:BBa_I746200-BBa_B0030-BBa_J23114_fw | 2013-09-06 | forward primer for BBa_I746200 with Gibson overhang for BBa_B0030-BBa_J23114 | ATGCTAGCTCTAGAGATTAAAGAGGAGAAATACTAG ATGAACAAGAAGATTCATTCCCTG |
| IK42:BBa_I746200_rev | 2013-09-06 | reverse primer for BBa_I746200 | TCAGAAGTGGGTGTTTACGCTC |
| IK43:pIK2-BBa_I746200_fw | 2013-09-06 | forward primer for pIK2 with Gibson overhang for BBa_I746200 | TGAGCGTAAACACCCACTTCTGA TACTAGAGTCACACTGGCTC |
| NK_01_FS_62_screening_L_BB(without_mRFP)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTCACCGACAAACAACAGATAAAACG |
| ygfG21C1 | 2013-06-03 | For insertion of methylmalonyl-CoA synthesis pathway into E. coli. Primer from <bib id="pmid17959404"/>. | TCACCGCGCACCGGCCTGCGGCAGCTCAGTGAATTTATCC AGATCTCGATCCC |
| ygfG21C2 | 2013-06-03 | For insertion of methylmalonyl-CoA synthesis pathway into E. coli. Primer from <bib id="pmid17959404"/>. | GTTATGCTGGATAATTTCTGCCGCTTCATTGGCGGTCATCC CAAAAAACCCCTCAAG |
Module Shuffling
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| AT01:RFC10prefix_TycA_fw | 2013-08-12 | Fw primer for amplification of TycAdCom; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG ATG TTA GCA AAT CAa GCC AAT C |
| AT02:RFC10suffix_TycA_rv | 2013-08-12 | Rv primer for amplification of TycAdCom; introduction of RFC10 suffix | TTTT CTGCAG CGGCCGC T ACTAGT A aGT TCG tTC TAC TTC TTT TTT C |
| AT03:RFC10prefix-TycB1_fw | 2013-08-12 | Fw primer for amplification of TycB1dCom; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG ATG AGT GTA TTT AGC AAA GAA CAA G |
| AT04:RFC10suffix_TycB1_rv | 2013-08-12 | Rv primer for amplification of TycB1dCom; introduction of RFC10 suffix | TTTT CTGCAG CGGCCGC T ACTAGT A TTC CTC CCC aCC TTC |
| AT05:RFC10prefix-TycC5_fw | 2013-08-12 | Fw primer for amplification of TycC5; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG AG GCG CAT ATT GCa GAG AG |
| AT06:RFC10suffix_TycC5_rv | 2013-08-14 | Rv primer for amplification of TycC5; introduction of RFC10 suffix | TTTT CTGCAG CGGCCGC T ACTAGT A TTT GGC TGT CTC TTC GAT GAA C |
| AT07:RFC10prefix-TycC6_fw | 2013-08-12 | Fw primer for amplification of TycC6; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG AG GGG AAT GTC TTC TCG ATC |
| AT08:RFC10suffix_TycC6_rv | 2013-08-14 | Rv primer for amplification of TycC6; introduction of RFC10 suffix | TTTT CTGCAG CGGCCGC T ACTAGT A TTA TTT CAG GAT aAA CAG TTC TTG |
| AT09:R10_B1_fw_longer | 2013-08-18 | Fw primer (longer) for amplification of TycB1dCom; introduction of RFC10 prefix | TTTT GAA TTC GCG GCC GCT TCT AG ATG AGT GTA TTT AGC AAA GAA CAA GTT C |
| AT10:R10_B1_rv_longer | 2013-08-18 | Rv primer (longer for amplification of TycB1dCom; introduction of RFC10 suffix | TTTT CTG CAG CGG CCG CTA CTA GTA ATA CGC aCT TTC CTC CCC GCC |
| AT11:R10_C5_fw_rpos | 2013-08-18 | Fw primer (repositioned) for amplification of TycC6; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG AG GAGCAGTTCGAGACGATCCAGCC |
| AT101 | 2013-09-05 | forward screening primer TycAdCom | GGACATCATCGAACAGGC |
| AT102 | 2013-09-05 | reverse screening primer TycAdCom | GACATAGGCAAGATCCGTAG |
| AT103 | 2013-09-05 | forward screening primer TycAdCom | GCAAGGACAAGTAGATGGC |
| AT104 | 2013-09-05 | reverse screening primer TycAdCom | GTACAGCCATACAGTCGC |
| AT105 | 2013-09-05 | forward screening primer TycC5 | GTCACCATGCTGAGAACG |
| AT106 | 2013-09-05 | reverse screening primer TycC5 | CAGCAAAGCGGTCATAATC |
| AT107 | 2013-09-05 | forward screening primer TycC6 | AAGCTACGCTGTTGATTGC |
| AT108 | 2013-09-05 | reverse screening primer TycC6 | AGCACGTAACGATCCTC |
| IK09:TycA_A1_fw | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycA A domain | ATGTTAGCAAATCAAGCCAATCTC |
| IK10:TycA_A1_rev | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycA A domain | TTGGTTTGCTGTAAGATCAGGCTC |
| IK11:TycB_A1_fw | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycB A1 domain | AATTCGGGAGTCGAGCTTTGTCAG |
| IK12:TycC6_rev | 2013-07-19 | reverse primer for tyrocidine TycC6 module | TTATTTCAGGATGAACAGTTCTTGCAGG |
| IK13:IK13:TycB1-dCom-dC_fw | 2013-07-19 | forward primer for tyrocidine TycB1 module without Com and C domain | GATTGCGTGGCAAACAATTCGGGAGTC |
| IK14:TycB1-TycC6_rev | 2013-07-19 | reverse primer for tyrocidine TycB1 module with Gibson overhang for TycC6 | CAGGCTCGATCGAGAAGACATTCCC TTCCTCCCCGCCTTCCACATACGC |
| IK15:TycC6-TycB1_fw | 2013-07-19 | forward primer for tyrocidine TycC6 module with Gibson overhang for TycB1 | GCGTATGTGGAAGGAGGGGAGGAA GGGAATGTCTTCTCGATCGAGCCTG |
| IK16:TycA_fw | 2013-07-19 | forward primer for tyrocidine TycA module | ATGTTAGCAAATCAGGCCAATCTCATC |
| IK17:TycA-dCom-TycC5_rev | 2013-07-19 | reverse primer for tyrocidine TycA module without Com domain with Gibson overhang for TycC5 | GAATGCGCTCTCGGCAATATGGGC TGTTCGCTCTACTTCTTTTTTCTCGG |
| IK18:TycC5-TycA-dCom_fw | 2013-07-19 | forward primer for tyrocidine TycC5 module with Gibson overhang for TycAdCom | CCGAGAAAAAAGAAGTAGAGCGAACA GCCCATATTGCCGAGAGCGCATTC |
| IK19:TycB1-TycC5_rev | 2013-07-19 | reverse primer for tyrocidine TycB1 module with Gibson overhang for TycC5 | GAATGCGCTCTCGGCAATATGGGC TTCCTCCCCGCCTTCCACATACGC |
| IK20:TycC5-TycB1_fw | 2013-07-19 | forward primer for tyrocidine TycC5 module with Gibson overhang for TycB1 | GCGTATGTGGAAGGAGGGGAGGAA GCCCATATTGCCGAGAGCGCATTC |
| IK21:pSB4K5-TycB1dComdC_rev | 2013-07-19 | reverse primer for mRFP-carrying backbones with Gibson overhang for tyrocidine TycA module (nomenclature wrong) | GTCGATGAGATTGGCCTGATTTGCTAACAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| IK22::pSB4K5-TycC6_fw | 2013-07-19 | forward primer for mRFP-carrying backbones with Gibson overhang for tyrocidine TycC6 module | AACATCCTGCAAGAACTGTTCATCCTGAAA TAATAACGCTGATAGTGCTAGTGTAGATC |
| IK23:pSB4K5-TycA_rev | 2013-07-19 | reverse primer for mRFP-carrying backbones with Gibson overhang for tyrocidine TycB1 module without Com and C domains + start codon (nomenklature wrong) | GACTCCCGAATTGTTTGCCACGCAATCCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW01:TycB_A1_rev | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycB A1 domain | CTTGGCACTTCCTTCAGGCTTC |
| PW02:TycB_E1_fw | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycB E domain | CGAGAGAGCGAGCAAGGTG |
| PW03:TycB_E1_rev | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycB E domain | GTACTCGCCTTCTTCTTTTGC |
| PW04:pSB1C3-TycC5ΔC_rev | 2013-07-19 | Integration of Tetrapeptide NRPS from Brevibacillus parabrevis in backbone; Gibson primer for Tetrapeptide I & II | TGTTTTGGTTGCGAGGAAGCTGTGCAGCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW05:TycC5ΔC_fwd | 2013-07-19 | Amplification of TycC5-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | ATGCTGCACAGCTTCCTCGCAACCAAAACAGCC |
| PW06:TycC5ΔC-TycB1ΔCom_rev | 2013-07-19 | Amplification of TycC5-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | GTGAAACAGCATCCCCTCTTGCATCGG AGGCTCGATCGAGAAGACATTCCCTTTG |
| PW07:TycC5ΔC-TycB1ΔCom_fwd | 2013-07-19 | Amplification of TycB1+C(TycB2)-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | CAAAGGGAATGTCTTCTCGATCGAGCCT CCGATGCAAGAGGGGATGCTGTTTCAC |
| PW08:C(TycB2)-TycAΔCom_rev | 2013-07-19 | Amplification of TycB1+C(TycB2)-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | GTTGTCGATGAGATTGGCCTGATTTGCTAACAT GATTTGCGCCAGCTCCTGCTCCGTGTT |
| PW09:C(TycB2)-TycAΔCom_fwd | 2013-07-19 | Amplification of TycA-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | AACACGGAGCAGGAGCTGGCGCAAATC ATGTTAGCAAATCAGGCCAATCTCATCGACAAC |
| PW10:TycAΔCom-TycC6_rev | 2013-07-19 | Amplification of TycAΔCom-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I | CTTTGGCTGTCTCTTCGATGAACGC TCGCTCTACTTCTTTTTTCTCGGTGCAATG |
| PW11:TycAΔCom-TycC6_fwd | 2013-07-19 | Amplification of TycC6-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I | CATTGCACCGAGAAAAAAGAAGTAGAGCGA GCGTTCATCGAAGAGACAGCCAAAG |
| PW12:TycAΔE-TycC6_rev | 2013-07-19 | Amplification of TycAΔE-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide II; one mismatch C->t | CTTTTGCACAGGCTCGATCGAGAAGAC GCTtTTGACAAAAAGAGCAACCTG |
| PW13:TycAΔE-TycC6_fwd | 2013-07-19 | Amplification of TycC6-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide II; one mismatch G->a | CAGGTTGCTCTTTTTGTCAAaAGC GTCTTCTCGATCGAGCCTGTGCAAAAG |
| PW14:C(TycC2)-indC_rev | 2013-08-16 | Amplification of C-domain from TycC2 from Brevibacillus parabrevis; Gibson overhang to IndC; for construct 1, 2 & 3 | ACATTGTGTAATATTATTTTCTAACAT CGTTTTGCTGCTGGCAGGCTG |
| PW15:C(TycC2)-indC_fwd | 2013-08-16 | Amplification of indC from Photorhabdus luminescens; Gibson overhang to C-domain from TycC2; for construct 1, 2 & 3 | CAGCCTGCCAGCAGCAAAACG ATGTTAGAAAATAATATTACACAATGT |
| PW16:indC_rev | 2013-08-16 | Amplification of indC from Photorhabdus luminescens; no Gibson overhang; for construct 1, 2 & 3 | TTAGATTATTTTCTCAATCTCAGCAACACCTTC |
| PW17:TycAdE-C(TycC2)_rev | 2013-08-16 | Amplification of TycAdE from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC2; for construct 1 | CGAAAGGAAGCGGGCCAGCTC AGCAACCTGCTCGATCGTCGGGTA |
| PW18:TycAdE-C(TycC2)_fwd | 2013-08-16 | Amplification of C-domain from TycC2-module from Brevibacillus parabrevis; Gibson overhang to TycAdE; for construct 1 | TACCCGACGATCGAGCAGGTTGCT GAGCTGGCCCGCTTCCTTTCG |
| PW19:TycC4dC_fwd | 2013-08-16 | Amplification of TycC4-module from Brevibacillus parabrevis without the C-domain; no Gibson overhang, ATG added; for construct 3 | ATGTATCCGCGCGATCTGACGATTC |
| PW20:TycC4dC-C(TycC2)_rev | 2013-08-16 | Amplification of TycC4-module from Brevibacillus parabrevis without the C-domain; Gibson overhang to C-domain form TycC2; for construct 3 | GGTGTACTCGGTTTTTTCCGA AATATGCGCAGCCAACTCATG |
| PW21:TycC4dC-C(TycC2)_fwd | 2013-08-16 | Amplification of C-domain from TycC2-module from Brevibacillus parabrevis; Gibson overhang to TycC4dC; for construct 3 | CATGAGTTGGCTGCGCATATT TCGGAAAAAACCGAGTACACC |
| PW22:pSB1C3-TycC4dC_rev | 2013-08-16 | Insertion of construct 3 into pSB1C3-backbone, amplification of pSB1C3; Gibson overhang to TycC4dC; for construct 3 | GAATCGTCAGATCGCGCGGATACAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW23:indC-pSB1C3_fwd | 2013-08-16 | Insertion of either fragments into pSB1C3-backbone, amplification of pSB1C3; Gibson overhang to indC from Photorhabdus luminescens; for constructs 1, 2 & 3 | GGTGTTGCTGAGATTGAGAAAATAATCTAA TAATAACGCTGATAGTGCTAGTGTAGATC |
| PW24:TycC1dC_fwd | 2013-08-16 | Amplification of the TycC1-module from Brevibacillus parabrevis without the C-domain; no Gibson overhang, ATG added; for construct 2 | ATGCAGACGAACAAACAACAGACG |
| PW25:pSB1C3-TycC1dC | 2013-08-16 | Insertion of construct 2 in pSB1C3-backbone, amplification of pSB1C3; Gibson overhang to TycC1-module without C-domain; for construct 2 | CGTCTGTTGTTTGTTCGTCTGCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW26:TycC5dC_fwd | 2013-09-04 | Amplification of TycC5-module without C-domain from Brevibacillus parabrevis; no Gibson overhang, ATG added; for constructs A & B | ATGCTGCACAGCTTCCTCGCAACC |
| PW27:TycC5dC-C(TycC4)_rev | 2013-09-04 | Amplification of TycC5-module without C-domain from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC4; for constructs A & B | CACATACGTCTCTTTTCCGCTCGT TTCGATGAACGCCGCCAGTTC |
| PW28:TycC5dC-C(TycC4)_fwd | 2013-09-04 | Amplification of C-domain from TycC4-module from Brevibacillus parabrevis; Gibson overhang to TycC5-module without C-domain; for constructs A & B | GAACTGGCGGCGTTCATCGAA ACGAGCGGAAAAGAGACGTATGTG |
| PW29:C(TycC4)-TycC4dC_rev | 2013-09-04 | Amplification of C-domain from TycC4-module from Brevibacillus parabrevis; Gibson overhang to TycC4-module without C-domain; for constructs A, B, C, E & G | GAATCGTCAGATCGCGCGGATA GGCAAACGTGTTGTTGAAATC |
| PW30:C(TycC4)-TycC4dC_fwd | 2013-09-04 | Amplification of TycC4-module without C-domain from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC4-module; for constructs A, B, C, E & G | GATTTCAACAACACGTTTGCC TATCCGCGCGATCTGACGATTC |
| PW31:TycC4-C(TycC4)_rev | 2013-09-04 | Amplification of TycC4-module from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC4-module; for constructs B & C | CACATACGTCTCTTTTCCGCTCGT GGCAATATGCGCAGCCAACTCATG |
| PW32:TycC4-C(TycC4)_fwd | 2013-09-04 | Amplification of C-domain from TycC4-module from Brevibacillus parabrevis; Gibson overhang to TycC4-module; for constructs B & C | CATGAGTTGGCTGCGCATATTGCC ACGAGCGGAAAAGAGACGTATGTG |
| PW33:TycC6dTE-C(TycC2)_rev | 2013-09-04 | Amplification of TycC6-module without the TE-domain from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC2-module; for constructs D & F | GGTGTACTCGGTTTTTTCCGA CGTGATGAAATCGGCCACCTTTTC |
| PW34:TycC6dTE-C(TycC2)_fwd | 2013-09-04 | Amplification of C-domain from TycC2-module from Brevibacillus parabrevis; Gibson overhang to TycC6-module without TE-domain; for constructs D & F | GAAAAGGTGGCCGATTTCATCACG TCGGAAAAAACCGAGTACACC |
| PW35:TycC6dTE-C(TycC4)_rev | 2013-09-04 | Amplification of TycC6-module without the TE-domain from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC4-module; for constructs E & G | CACATACGTCTCTTTTCCGCTCGT CGTGATGAAATCGGCCACCTTTTC |
| PW36:TycC6dTE-C(TycC4)_fwd | 2013-09-04 | Amplification of C-domain from TycC4-module from Brevibacillus parabrevis; Gibson overhang to TycC6-module without TE-domain; for constructs E & G | GAAAAGGTGGCCGATTTCATCACG ACGAGCGGAAAAGAGACGTATGTG |
| PW37:pSB1C3-TycC5dC_rev | 2013-09-04 | Insertion of constructs A & B in pSB1C3-backbone, amplification of pSB1C3; Gibson overhang to TycC5-module without C-domain; for constructs A & B | GGTTGCGAGGAAGCTGTGCAGCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
Linker Variation
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| AR01 | 2013-09-17 | sTycC4dC_fw | ATG AAGCACTTGCTGCCGCTCGTC |
| AR02 | 2013-09-17 | c(TycC2)s_rev | TTGTGTAATATTATTTTCTAACAT CGCTACCTGTTGCAACCGCTC |
| AR03 | 2013-09-17 | s-Ind_fw | GAGCGGTTGCAACAGGTAGCG ATGTTAGAAAATAATATTACAATGT |
| AR04 | 2013-09-17 | c(TycC2)l_rev_l | TTGTGTAATATTATTTTCTAACAT AAGCGACTGCTGGTTCGCG |
| AR05 | 2013-09-17 | l-Ind_fw | AACGCGAACCAGCAGTCGCTT ATGTTAGAAAATAATATTACACAATGT |
| AR06 | 2013-09-17 | lTycC4dC_fw | ATG CGCAACTATCCGGTCGAGACG |
| AR07 | 2013-09-17 | TycC4dCom_short-BB_rev | GACGAGCGGCAGCAAGTGCTTCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| AR08 | 2013-09-17 | TycC4dCom_long-BB_rev | CGTCTCGACCGGATAGTTGCGCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW19:TycC4dC_fwd | 2013-08-16 | Amplification of TycC4-module from Brevibacillus parabrevis without the C-domain; no Gibson overhang, ATG added; for construct 3 | ATGTATCCGCGCGATCTGACGATTC |
| PW20:TycC4dC-C(TycC2)_rev | 2013-08-16 | Amplification of TycC4-module from Brevibacillus parabrevis without the C-domain; Gibson overhang to C-domain form TycC2; for construct 3 | GGTGTACTCGGTTTTTTCCGA AATATGCGCAGCCAACTCATG |
| PW21:TycC4dC-C(TycC2)_fwd | 2013-08-16 | Amplification of C-domain from TycC2-module from Brevibacillus parabrevis; Gibson overhang to TycC4dC; for construct 3 | CATGAGTTGGCTGCGCATATT TCGGAAAAAACCGAGTACACC |
| PW14:C(TycC2)-indC_rev | 2013-08-16 | Amplification of C-domain from TycC2 from Brevibacillus parabrevis; Gibson overhang to IndC; for construct 1, 2 & 3 | ACATTGTGTAATATTATTTTCTAACAT CGTTTTGCTGCTGGCAGGCTG |
Domain Shuffling and PPTases
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| KH1_ccdb_fw | 2013-07-23 | amplifing ccdb with RBS from pDONR | TGGTGCCATTACATACAGATACT GAGAACAGGGGCTGGTGAAATGC |
| KH2_ccdb_rv | 2013-07-23 | amplifing ccdb with RBS from pDONR | AGAGTCTGTCTGTTCAATCCACTT TTATATTCCCCAGAACATCAGGTTAATGGCG |
| KH3_indC_backbone-withoutT_fw | 2013-07-23 | linearization of vector with partial indigoidine synthase | AAGTGGATTGAACAGACAGACTCTAAAAC |
| KH4_indC_backbone-withoutT_rv | 2013-07-23 | linearization of vector with partial indigoidine synthase | AGTATCTGTATGTAATGGCACCAATAGACGC |
| KH5_indC_T_fw | 2013-07-23 | introducing T domain of indC | TGGTGCCATTACATACAGATACT GAAATAAGGCTTGGAAAAATTTGGATGGAAGT |
| KH6_indC_T_rv | 2013-07-23 | introducing T domain of indC | AGAGTCTGTCTGTTCAATCCACTT AGCCAATTCTGCTATATTAGGAGATTGA |
| KH7_bpsA_T_fw | 2013-07-23 | introducing T domain of bpsA | TGGTGCCATTACATACAGATACT GAGAAGGAAATCGCAGCCGTGTGGG |
| KH8_bpsA_T_rv | 2013-07-23 | introducing T domain of bpsA | AGAGTCTGTCTGTTCAATCCACTT GGCCAGCTTTTCAATTGTTGGTGACTCC |
| KH9_ccdB-Big_fw | 2013-08-06 | ccdB_Big amplified from pDONOR (Dominik) for indC(ccdB) construct | TGCCATTACATACAGATACT ACTGGCTGTGTATAAGGGAGCCTGAC |
| KH10_ccdB-Big_rv | 2013-08-06 | ccdB_Big amplified from pDONOR (Dominik) for indC(ccdB) construct | AGAGTCTGTCTGTTCAATCCACTT CGCGTGGATCCGGCTTAC |
| KH11_plu2642-T_fw | 2013-08-14 | T domain exchange plu2642 | TGGTGCCATTACATACAGATACT GAAACACAGATCGTAAAGATATGG |
| KH12_plu2642-T_rv | 2013-08-14 | T domain exchange plu2642 | AGAGTCTGTCTGTTCAATCCACTT TGCCAATTGTTTAACCGTTG |
| KH13_BBa-entD_fw | 2013-08-09 | submitting endD to registry | GCAT GAATTCGCGGCCGCTTCTAG ATGAAAACTACGCATACCTCCCTC |
| KH14_BBa-entD_rv | 2013-08-09 | submitting endD to registry | GCAT CTGCAGCGGCCGCTACTAGTA TCATTAATCGTGTTGGCACAGCG |
| KH15_BBa-sfp_Bsub_fw | 2013-08-09 | submitting sfp_Bsub to registry | GCAT GAATTCGCGGCCGCTTCTAG ATGAAGATTTACGGAATTTATATGGAC |
| KH16_BBa-sfp_Bsub_rv | 2013-08-09 | submitting sfp_Bsub to registry | GCAT CTGCAGCGGCCGCTACTAGTA TCATTATAAAAGCTCTTCGTACGAGAC |
| KH17_BBa-svp_Svert_fw | 2013-08-09 | submitting svp_Svert to registry | GCAT GAATTCGCGGCCGCTTCTAG A TGATCGCCGCCCTCCTGCCCTCC |
| KH18_BBa-svp_Svert_rv | 2013-08-09 | submitting svp_Svert to registry | GCAT CTGCAGCGGCCGCTACTAGTA AGCTAGCTCATTACGGGAC |
| KH19_BBa-delC_fw | 2013-08-09 | submitting delC to registry | GCAT GAATTCGCGGCCGCTTCTAG ATGCAGCTCGTGTCCGTGCG |
| KH20_BBa-delC_rv | 2013-08-09 | submitting delC to registry | GCAT CTGCAGCGGCCGCTACTAGTA TTA TCATGTCGATTCCTTGGTGC |
| KH21_BBa-indC_fw | 2013-08-09 | submitting indC to registry | GCAT GAATTCGCGGCCGCTTCTAG ATGTTAGAAAATAATATTACACAATG |
| KH22_BBa-indC_rv | 2013-08-09 | submitting indC to registry | GCAT CTGCAGCGGCCGCTACTAGTA TTAGATTATTTTCTCAATCTCAG |
| NI01:bpsA_AOxA_PstI_fw | 2013-06-04 | for amplifying bpsA (AOxA domain) | GAGGAGAAATACTAGATGACACTGCAGGAAACAAGCGTGC |
| NI02:bpsA_AOxA_rv | 2013-06-04 | for amplifying bpsA (AOxA domain) | GAAGGGCCGTTCCACCAGCTCAGCGTTGACCTGGTCAGAAGCG |
| NI03:bpsA_T_fw | 2013-06-04 | for amplifying bpsA (T domain) | GACCAGGTCAACGCTGAGCTGGTGGAACGGCCCTTCGTCG |
| NI04:bpsA_T_rv | 2013-06-04 | for amplifying bpsA (T domain) | GACGAAGCGACTAGACTCCTGAGCCACTTCTCTCTCCAGCCG |
| NI05:bpsA_TE_fw | 2013-06-04 | for amplifying bpsA (TE domain) with RBS 1 | GAGAGAGAAGTGGCTCAGGAGTCTAGTCGCTTCGTCCGA |
| NI06:bpsA_TE_Stop_XbaI_rbs_rv | 2013-06-04 | for amplifying bpsA (TE domain) with RBS 1 | TGGCAGCAGAGCAGCGATCATCTAGTATTT CTCCTCTTTCCTCTAGATCATCATTCCC CCAGCAGGTATCTAAT |
| NI07:svp_fw | 2013-06-04 | for amplifying svp | GAGGAGAAATACTAGATGATCGCTGCTCTGCTGCCAAGTTGG |
| NI08:svp_rv | 2013-06-04 | for amplifying svp | GCACTATCAGCGTTATTATGGCACGGCAGTCCTATCGTCG |
| NI09:pSB1C3_fw | 2013-06-04 | for linearizing, amplifying pSB1C3 | GATAGGACTGCCGTGCCATAATAA CGCTGATAGTGCTAGTGTAGATCGC |
| NI10:pSB1C3_PstI_rv | 2013-06-04 | for linearizing, amplifying pSB1C3 | GCTTGTTTCCTGCAGTGTCATCTAG TATTTCTCCTCTTTCTCTAGTATGTG |
| NI11:bpsA_rvN | 2013-07-15 | for amplifying bpsA (TE domain) with RBS 2 | TGGCAGCAGAGCAGCGATCATTATTTAGGTTTCCTGTGTGAATCATCATTCCCCCAGCAGGTATCTAAT |
| NI12:svp_fwN | 2013-07-15 | for amplifying svp with RBS 2 | CAGGAAACCTAAATAATGATC GCTGCTCTGCTGCCAAGTTGG |
| NI13_ccdb_fw | 2013-07-23 | amplifing ccdb with RBS from pDONR | CGTCGCACCTAGGACCGAAACA GAGAACAGGGGCTGGTGAAATGC |
| NI14_ccdb_rv | 2013-07-23 | amplifing ccdb with RBS from pDONR | GCCACTTCTCTCTCCAGCCGTCG TTATATTCCCCAGAACATCAGGTTAATGGCG |
| NI15_bpsA_backbone-withoutT_fw | 2013-07-23 | linearization of vector with partial indigoidine synthase | CGACGGCTGGAGAGAGAAGTGGC |
| NI16_bpsA_backbone-withoutT_rv | 2013-07-23 | linearization of vector with partial indigoidine synthase | TC TGTTTCGGTCCTAGGTGCGACG |
| NI17_indC_T_fw | 2013-07-23 | introducing T domain of indC | CGTCGCACCTAGGACCGAAACA GAAATAAGGCTTGGAAAAATTTGGATGGAAGT |
| NI18_indC_T_rv | 2013-07-23 | introducing T domain of indC | GCCACTTCTCTCTCCAGCCGTCG AGCCAATTCTGCTATATTAGGAGATTGA |
| NI19_bpsA_T_fw | 2013-07-23 | introducing T domain of bpsA | CGTCGCACCTAGGACCGAAACA GAGAAGGAAATCGCAGCCGTGTGGG |
| NI20_bpsA_T_rv | 2013-07-23 | introducing T domain of bpsA | GCCACTTCTCTCTCCAGCCGTCG GGCCAGCTTTTCAATTGTTGGTGACTCC |
| NI19:ngrA_fw | GAATTCGCGGCCGCTTCTAG ATGGAACAGACAGTTATACACACC | ||
| NI20:ngrA_rv | CTGCAGCGGCCGCTACTAGTA TTATTGAATAATTAGCGTTAATATTTCCTG | ||
| RB05:RBS1-BpsA_fw | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | AAAGAGGAGAAA TACTAG ATGACACTTCAGGAAACCAGCG |
| RB06:BpsA-RBS2_rv | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | CTTTCCTGTGTGA AAGCTT TCA TCACTCCCCGAGCAGATATCG |
| RB07:RBS2-svp_fw | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | CTTTCACACAGGAAAG TAAATA ATGGCTGCTCTTCTTCCTAGTTGGGC |
| RB08:svp-pSB1C3_rv | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | CTATCAGCGTTATTA AAGCTT TCATCA TGGCACGGCAGTCCTATCG |
| RB09:pSB1C3_fw | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | AAGCTT TAATAACGCTGATAGTGCTAGTGTAGATCGC |
| RB10:pSB1C3-RBS1_rv | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | CTAGTA TTTCTCCTCTTT CTCTAGTATGTGTG |
| RB11:BpsA_Taka_fw | 2013-07-08 | colony PCR bpsA Takahashi | CAT ATGACTCTTCAGGAGACCAGCGTGCTC |
| RB12:BpsA_Taka_rv | 2013-07-08 | colony PCR bpsA Takahashi | AAG CTTCTCGCCGAGCAGGTAGCGGATGTG |
| RB13:svp_Taka_fw | 2013-07-08 | colony PCR svp Takahashi | CAT ATGATCGCCGCCCTCCTGCCCTCCTG |
| RB14:svp_Taka_rv | 2013-07-08 | colony PCR svp Takahashi | CTCGAGCGGGACGGCGGTCCGGTCGTCCGC |
| RB15:svp_Sanch_fw | 2013-07-08 | PCR svp Sanchez | TATA ATGCATGCTCGCCGCCCTCCCC |
| RB16:svp_Sanch_rv | 2013-07-08 | PCR svp Sanchez | TTAAGATCTCGGGACGGCGGTCCGGTC |
| RB17:sfp_fw | 2013-07-08 | sfp extraction from BAP1 | ATGAAGATTTACGGAATTTATATGG |
| RB18:sfp_rv | 2013-07-08 | sfp extraction from BAP1 | TTATAAAAGCTCTTCGTACGAGACC |
| RB19:entD_fw | 2013-07-08 | entD extraction from E.coli Lambalot | TAAATA ATGGTCGATATGAAAACTACGC |
| RB20:entD_rv | 2013-07-08 | entD extraction from E.coli Lambalot | AAGCTT ATTAATCGTGTTGGCACAGCG |
| RB21:pSB1C3_fw | 2013-07-15 | indigoidine exchangeable construct | TAATGA GCTAGC TAATAACGCTGATAGTGCTAGTG |
| RB22:pSB1C3_rv | 2013-07-15 | indigoidine exchangeable construct | CAT GGTACC TTTCTCCTCTTT CTCTAGTATGTGTG |
| RB23:bpsA-fus_fw | 2013-07-15 | indigoidine exchangeable construct | CACACATACTAGAG AAAGAGGAGAAA GGTACC ATGACTAGTACACTGCAGG |
| RB24:bpsA-fus_rv | 2013-07-15 | indigoidine exchangeable construct | CAT GGATCC GGTTTCCTGTGTGAA TCATTA TTCCCCCAGCAGGTATCTAATATG |
| RB25:svp-fus_fw | 2013-07-15 | indigoidine exchangeable construct | CACAGGAAACC GGATCC ATGACTAGTATCGCTGCTCTGCTG |
| RB26:svp-fus_rv | 2013-07-15 | indigoidine exchangeable construct | CAGCGTTATTA GCTAGC TCATTA TGGCACGGCAGTCCTATC |
| RB27:indC-Plum_fw | 2013-07-15 | indigoidine exchangeable construct | CATACTAGAG AAAGAGGAGAAA GGTACC ATGTTAGAAAATAATATTACACAATG |
| RB28:indC-Plum_rv | 2013-07-15 | indigoidine exchangeable construct | CAT GGATCC GGTTTCCTGTGTGAA TTA TTAGATTATTTTCTCAATCTCAG |
| RB29:svp-Svert_fw | 2013-07-15 | indigoidine exchangeable construct | CACAGGAAACC GGATCC A TGATCGCCGCCCTC |
| RB30:svp-Svert_rv | 2013-07-15 | indigoidine exchangeable construct | CAGCGTTATTA GCTAGC TCA TTACGGGACGGCGGTC |
| RB31:Sc-indC-Schr_fw | 2013-07-15 | indigoidine exchangeable construct | GAG AAAGAGGAGAAA GGTACC ATGAGCGTAGAGACCATC |
| RB32:Sc-indC-Schr_rv | 2013-07-15 | indigoidine exchangeable construct | CAT AGATCT GGTTTCCTGTGTGAA TTA TCAGTAGTTGGGCGTCTTG |
| RB33:entD-MG1655_fw | 2013-07-15 | indigoidine exchangeable construct | CACAGGAAACC GGATCC ATGAAAACTACGCATACCTC |
| RB34:entD-MG1655_rv | 2013-07-15 | indigoidine exchangeable construct | CAGCGTTATTA GCTAGC TCA TTAATCGTGTTGGCACAGC |
| RB35:sfp-Naka_fw | 2013-07-15 | indigoidine exchangeable construct | CACAGGAAACC GGATCC ATGAAGATTTACGGAATTTATATGGAC |
| RB36:sfp-Naka_rv | 2013-07-15 | indigoidine exchangeable construct | CAGCGTTATTA GCTAGC TCA TTATAAAAGCTCTTCGTACGAGAC |
| RB37:Plum-extr_fw | 2013-07-15 | genomic extraction | ATGTTAGAAAATAATATTACACAATG |
| RB38:Plum-extr_rv | 2013-07-15 | genomic extraction | TTAGATTATTTTCTCAATCTCAG |
| RB39:Svert-extr_fw | 2013-07-15 | genomic extraction | GTGATCGCCGCCCTC |
| RB40:Svert-extr_rv | 2013-07-15 | genomic extraction | TTACGGGACGGCGGTC |
| RB41:MG1655-extr_fw | 2013-07-15 | genomic extraction | ATGAAAACTACGCATACCTCCCTC |
| RB42:MG1655-extr_rv | 2013-07-15 | genomic extraction | TTAATCGTGTTGGCACAGCGTTATG |
| RB43:Bsub-extr_fw | 2013-07-15 | genomic extraction | ATGAAGATTTACGGAATTTATATGGAC |
| RB44:Bsub-extr_rv | 2013-07-15 | genomic extraction | TTATAAAAGCTCTTCGTACGAGAC |
| RB45:Sc-indC-Schr_rvG | 2013-07-15 | indigoidine exchangeable construct | CAT GGATCC GGTTTCCTGTGTGAA TTA TCAGTAGTTGGGCGTCTTG |
| RB46:indC-woPPT_rv | 2013-08-05 | indC-pSB1C3 w/o PPTase; RB21 overlap | GCACTATCAGCGTTATTA GCTAGCTCATTA TTA TTAGATTATTTTCTCAATCTCAG |
| RB47:indC-SpeI_fw | 2013-08-05 | remove cutting sites from indC-Plum | CAAGTTCCTAAACCC ACTAGT CTGGCTTAT ATTATTTATACCTCTGGTAGCAC |
| RB48:indC-EcoRI_rv | 2013-08-05 | remove cutting sites from indC-Plum | CAATACCCACC GAATTC TTTGAGCT AATTTCTGACAGACAATACC |
| RB49:indC-EcoRI_fw | 2013-08-05 | remove cutting sites from indC-Plum | AGCTCAAA GAATTC GGTGGGTATTG GGCTTTTTTGTGATC |
| RB50:indC-SpeI_rv | 2013-08-05 | remove cutting sites from indC-Plum | ATAAGCCAG ACTAGT GGGTTTAGGAACTTG GAACTTGAACTGTG |
| RB51:pSB(2/3)K3_fw | 2013-08-05 | PPTase plasmid | TAATGA GCTAGC tactagtagcggccgctgcagtc |
| RB52:pSB(2/3)K3_rv | 2013-08-05 | PPTase plasmid | CAT GGATCC GGTTTCCTGTGTGAA ctctagaagcggccgcgaattcc |
| RB53:entF-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAAACGATTATCGCCGCGGCATTC |
| RB54:entF-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT TGCCAGTTTGGCGACAGTTGACG |
| RB55:tycA1-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAATCGATTCTCGTCTCCATCTGG |
| RB56:tycA1-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT AGCAACCTGCTCGATCGTCGGGTAATTC |
| RB57:tycC6-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAACAGCAACTGGCAGCCATCTGGCAAG |
| RB58:tycC6-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT GGCCACCTTTTCGATCGTCGGATACTG |
| RB59:delH4-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAGACGCTGCTGGCCCGTATCTG |
| RB60:delH4-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT CGCCAGCTCGGCAATGCTTTGC |
| RB61:delH5-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GCCATGGCGCTGGCCCGCATC |
| RB62:delH5-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT CAGCAGGCCGGCAATGGTCG |
| RB63:pSB(2/3)K3_rv_korr | 2013-08-07 | PPTase plasmid | CAT GGATCC GGTTTCCTGTGTGAA CTCTAGTATGTGTGAAATTGTTATCC |
| RB64:DelC-extr_fw | 2013-08-07 | get Delftia PPTase | ATGCAGCTCGTGTCCGTGCGTGAG |
| RB65:DelC-extr_rv | 2013-08-07 | get Delftia PPTase | TCATGTCGATTCCTTGGTGCGCCAC |
| RB66:DelC-pSB2K3_fw | 2013-08-07 | DelC in PPTase-construct | CACAGGAAACC GGATCC ATGCAGCTCGTGTCCGTGCGTGAG |
| RB67:DelC-pSB2K3_rv | 2013-08-07 | DelC in PPTase-construct | CAGCGTTATTA GCTAGC TCA TCATGTCGATTCCTTGGTGCGCCAC |
| RB68:indC-SpeI_fw_corr | 2013-08-09 | indC w/o cutting sites corrected | CAAGTTCCTAAACCC ACgAGT CTGGCTTAT ATTATTTATACCTCTGGTAGCAC |
| RB69:indC-EcoRI_rv_corr | 2013-08-09 | indC w/o cutting sites corrected | CAATACCCACC GAAcTC TTTGAGCT AATTTCTGACAGACAATACC |
| RB70:indC-EcoRI_fw_corr | 2013-08-09 | indC w/o cutting sites corrected | AGCTCAAA GAgTTC GGTGGGTATTG GGCTTTTTTGTGATC |
| RB71:indC-SpeI_rv_corr | 2013-08-09 | indC w/o cutting sites corrected | ATAAGCCAG ACTcGT GGGTTTAGGAACTTG GAACTTGAACTGTG |
| RB72:bpsA-TTE_rv | 2013-08-09 | exchange T and TE-domain | GCACTATCAGCGTTATTA GCTAGC TCATTA TTCCCCCAGCAGGTATCTAATATG |
| RB73:entF-TTE_rv | 2013-08-09 | exchange T and TE-domain | GCACTATCAGCGTTATTA GCTAGC TCA TTACCTGTTTAGCGTTGCGCGAATAATCG |
| RB74:tycC6-TTE_rv | 2013-08-09 | exchange T and TE-domain | GCACTATCAGCGTTATTA GCTAGC TCA TTATTTCAGGATGAACAGTTCTTGC |
| RB75:delH5-TTE_rv | 2013-08-09 | exchange T and TE-domain | GCACTATCAGCGTTATTA GCTAGC TCATTA TCAGTCCAGCGCGTACTCCAG |
| RB76:plu2670-T_fw | 2013-08-15 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAAACCACACTGGCTGCTATC |
| RB77:plu2670-T_rv | 2013-08-15 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT CGCAAATGCGGATAACACC |
| RB78:indC-SpeI_rv_corr2 | 2013-08-26 | indC w/o cutting sites w/o frameshift | ATAAGCCAG ACTcGT GGGTTTAGGAACTTG AACTGTG |
| RB79:indC_woPPTase_rv2 | 2013-08-29 | indC-pSB1C3 w/o PPTase; RB35 5' part | CAGCGTTATTA GCTAGC TCA TTAGATTATTTTCTCAATCTCAG |
| RB80:indCdT_3_fw | 2013-09-06 | GGCGGATATCCTATGAGTTTGAGATTGC | |
| RB81:indCdT_3_rv | 2013-09-06 | TCCATTGGCCGTCAAAGGTAATTTATCG | |
| RB82:bpsA-T3_fw | 2013-09-06 | CGATAAATTACCTTTGACGGCCAATGGA AAGATCGATGTGAAAGCACTGGCCGCTTCTGACC | |
| RB83:bpsA-T3_rv | 2013-09-06 | GCAATCTCAAACTCATAGGATATCCGCC CAGACCTGGCCAGCAGATCACAGG | |
| RB84:indCdT_4_fw | 2013-09-06 | TTTGAAGTTGCATACCAGCTTGAACAAGC | |
| RB85:bpsA-T4_rv | 2013-09-06 | GCTTGTTCAAGCTGGTATGCAACTTCAAA TGCCACGCGAGCTCCGAAGCTATATCC | |
| RB86:indC-T2_fw | 2013-09-06 | TTACAGATACTTTTTCAATCTCCTAATATAGCAG | |
| RB87:indC-T2_rv | 2013-09-06 | TACTTCCATCCAAATTTTTCCAAGC | |
| RB88:bpsA-T2_fw | 2013-09-06 | GCTTGGAAAAATTTGGATGGAAGTA CTGAGACGCGAAAATGCTAGTGTCC | |
| RB89:bpsA-T2_rv | 2013-09-06 | TTAGGAGATTGAAAAAGTATCTGTAA AGGCAGGGACACTCCCAGTCTAGCATTCAG | |
| RB90:indC-T-screen_fw | 2013-09-13 | GATCAATGCGGCCTTTAATATTCG | |
| RB91:bpsA-T-screen_fw | 2013-09-13 | GGAACTGAATGCTAGACTGGGAG | |
| RB92:entF-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGCTGGATCGCAAAGCCTTACCGTTG | |
| RB93:tycA1-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGATCGACCGCAAAGCGTTGC | |
| RB94:tycC6-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAAGTGGATCGCAAAGCTTTG | |
| RB95:delH4-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGCTGGACCGGCAGGCCCTG | |
| RB96:delH5-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGCTTGACCGTGGTGCGCTG | |
| RB97:plu2642-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAAATCGATTTCGACACATTACAAG | |
| RB98:plu2670-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGCTGGACCGTCGGGCGTTACCGGCAC | |
| RB99:plu2642_pRB23-plu_fw | 2013-09-24 | plu2642 on pSB1C3 | CATACTAGAG AAAGAGGAGAAA GGTACC ATGCAATCAACTCTCCCAATAATAAAATGG |
| RB100:plu2642_pRB23-plu_rv | 2013-09-24 | plu2642 on pSB1C3 | CAGCGTTATTA GCTAGC TCA TCAACTCAAGAATCGAGCTAATTCGTTAACAC |
| RB101:plu2642_A_rv | 2013-09-24 | GTTCTGGCATCCTTCTTAATTAACAT TGCAGCGTGTTTCACTTGATCGATAAC | |
| RB102:indC_Ox_fw | 2013-09-24 | ATGTTAATTAAGAAGGATGCCAGAAC | |
| RB103:plu2642_valInd_fw | 2013-09-24 | CAGCCTGCCAGCAGCAAAACG ATGCAATCAACTCTCCCAATAATAAAATGG | |
| RB104:plu2642_valInd_rv | 2013-09-24 | CAT CGTTTTGCTGCTGGCAGGCTG | |
| RB105:tycC2_T3_fw | 2013-09-24 | CGATAAATTACCTTTGACGGCCAATGGA AAAGTGGATCGCAAGGCATTG | |
| RB106:tycC2_T1_rv | 2013-09-24 | AGAGTCTGTCTGTTCAATCCACTT GGCCAGGCCCGCGATCGTTGG | |
| RB107:tycC2_A_fw | 2013-09-24 | CATACTAGAG AAAGAGGAGAAA GGTACC ATGACGGAAGCGGAAAAACGCACACTCCTTC | |
| RB108:tycC2_A_rv | 2013-09-24 | GTTCTGGCATCCTTCTTAATTAACAT GACGACCGCTTTGACCGCTTCATG | |
| RB109:tycC2_valInd_fw | 2013-09-24 | CAGCCTGCCAGCAGCAAAACG ATGACGGAAGCGGAAAAACGCACACTCCTTC | |
| RB110:plu2642_TE_fw | AGTGGATTGAACAGACAGACTCT AAAATCGTTGAAGGTGAAGTAACCAG |
Plasmids
Delftibactin
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
| pIK1 | 2013-08-12 | Methylmalonyl-CoA pathway with permeability device and sfp | pSB3C5-lacP-BBa_B0034-BBa_I746200-BBa_B0029-pcc-acca2-BBa_B0030-sfp | 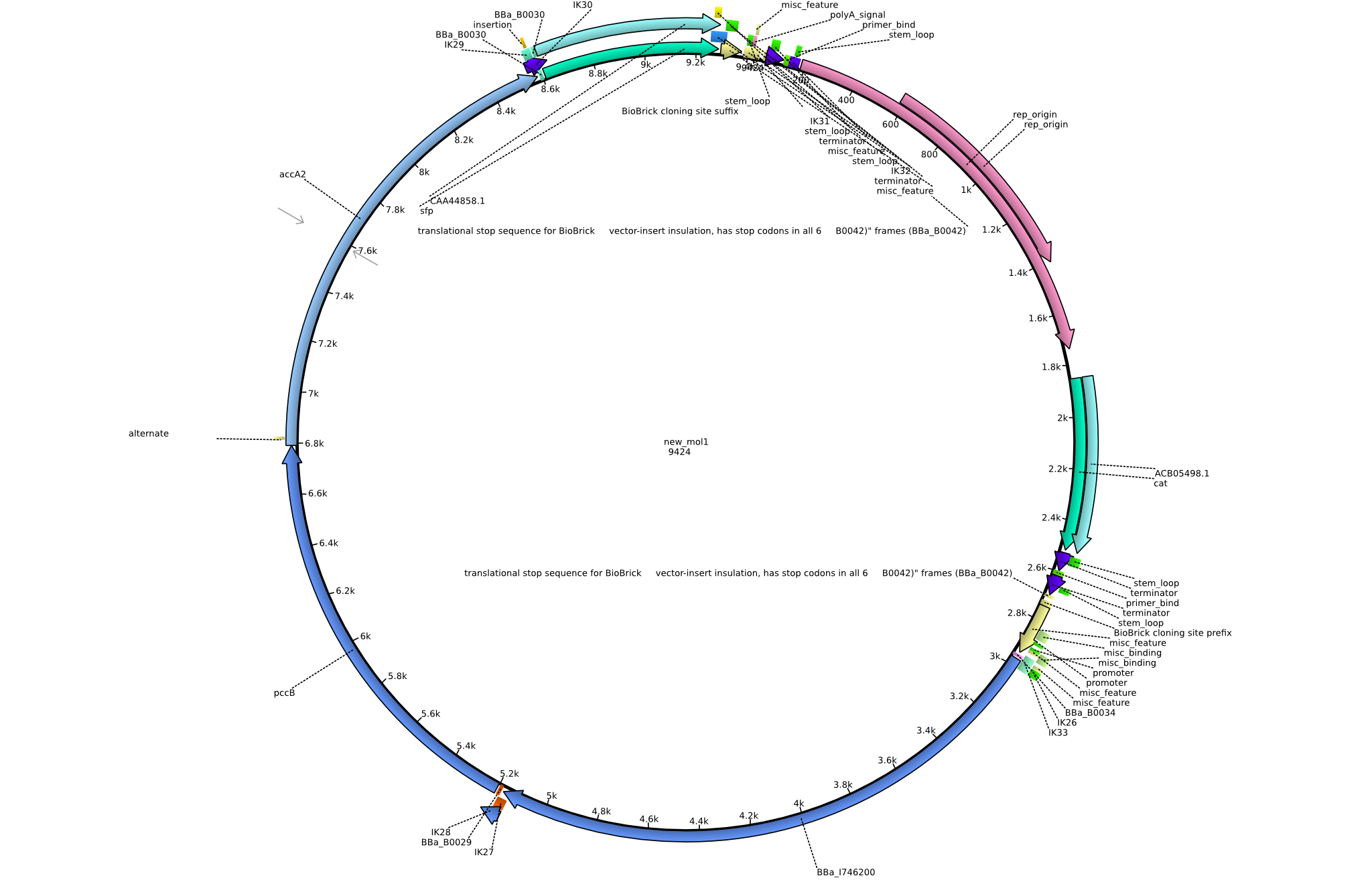 | pIK1 |
| pIK2 | 2013-08-12 | Methylmalonyl-CoA pathway with sfp | pSB3C5-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp | 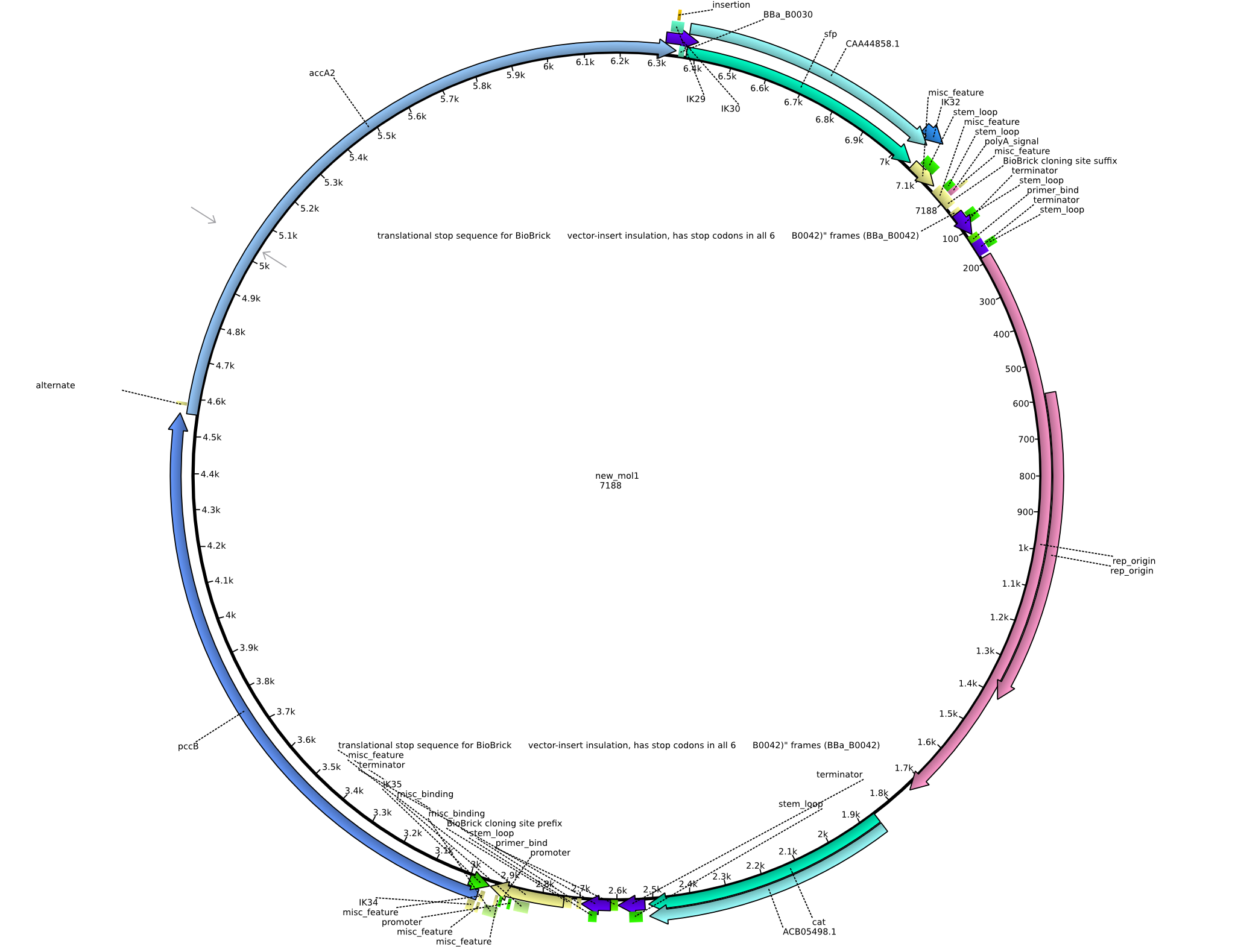 | pIK2 |
| pIK7 | 2013-08-28 | Methylmalonyl-CoA pathway with sfp | pSB3K3-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp | 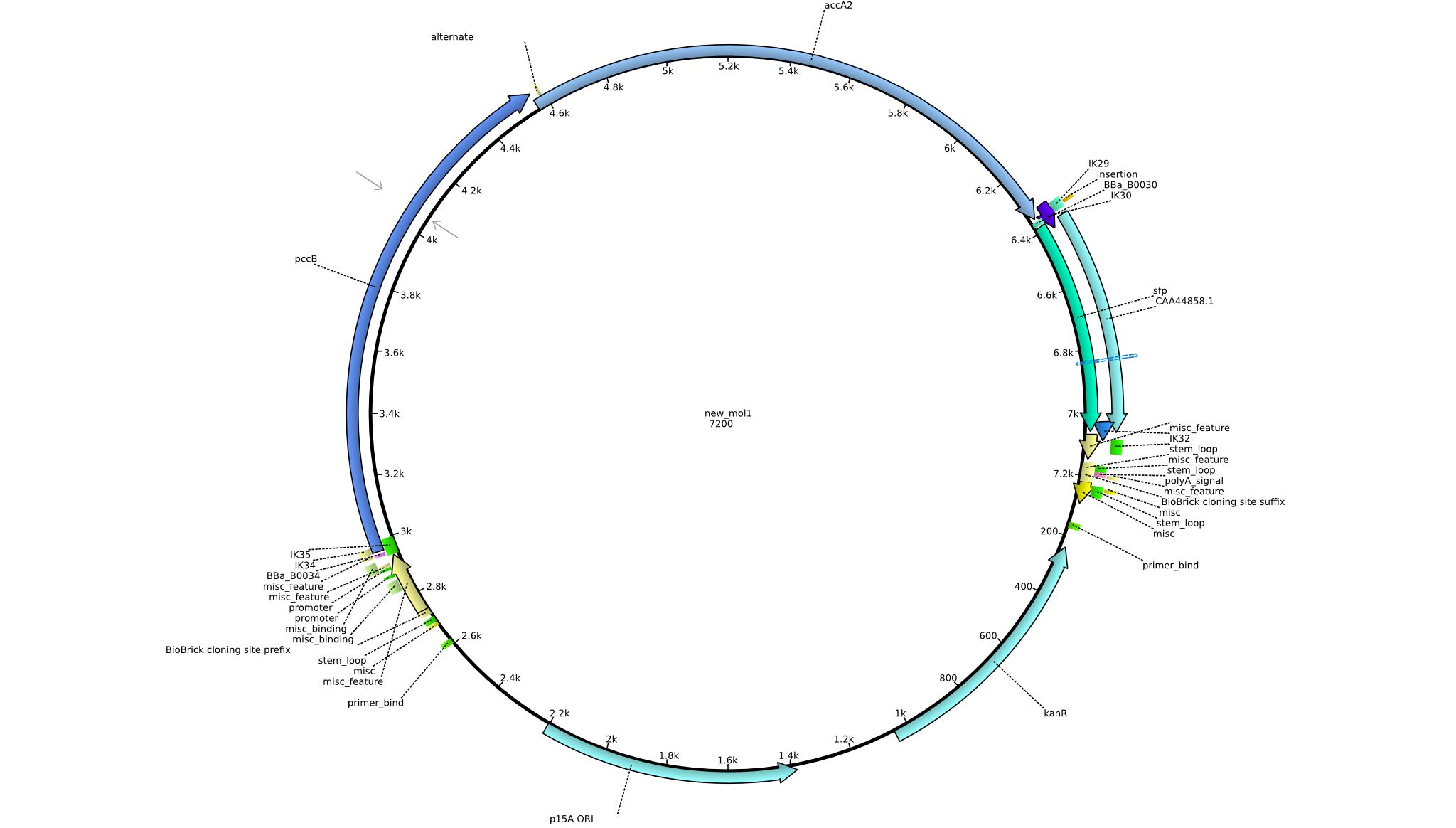 | pIK7 |
| pIK8 | 2013-09-12 | Methylmalonyl-CoA pathway with sfp and the permeability device behind a weak promoter | pSB3C5-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp-BBa_B0010-BBa_B0030-BBa_I746200-BBa_B0012 | 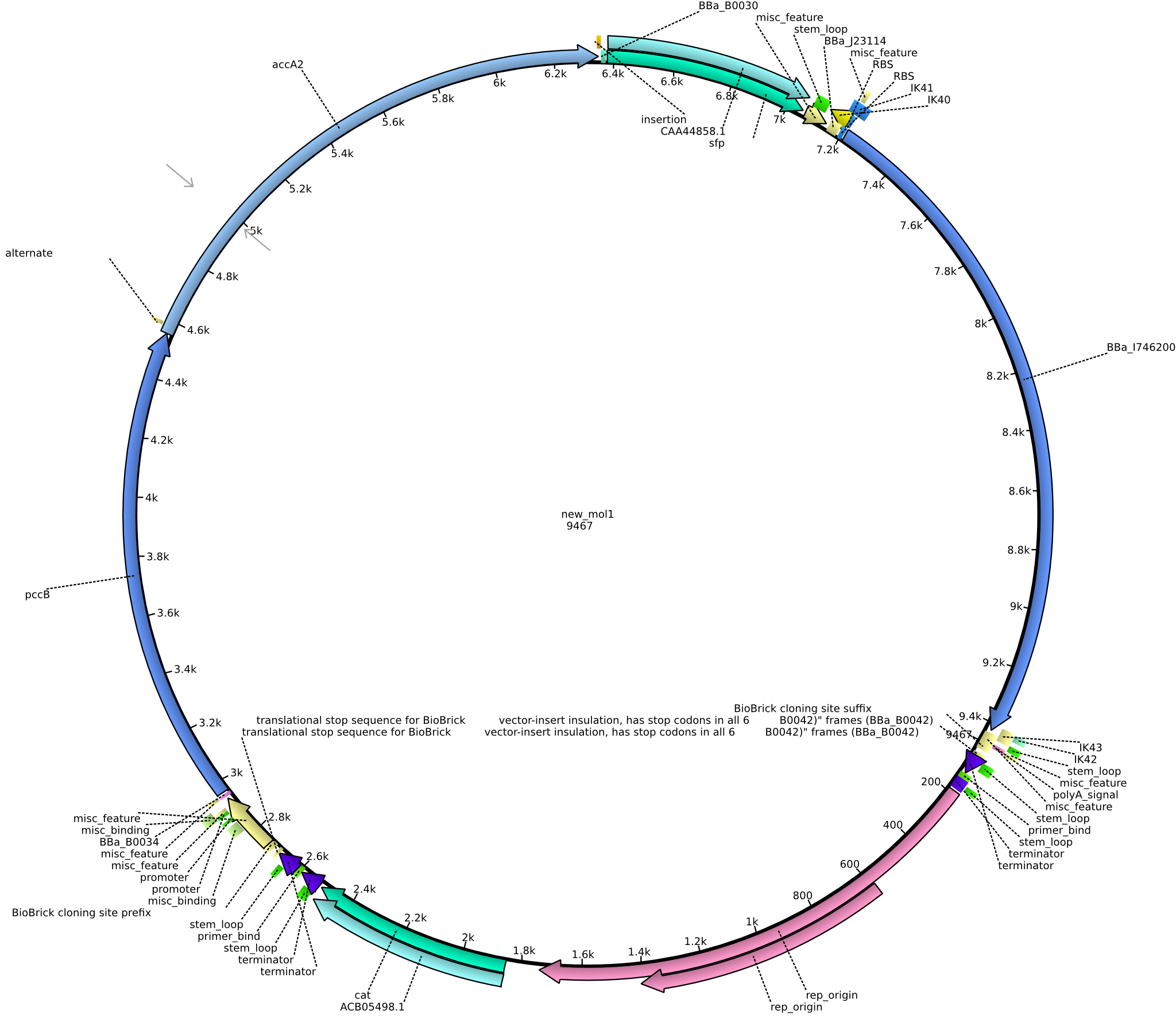 | pIK8 |
| pIK9 | 2013-09-18 | Methylmalonyl-CoA pathway with sfp and the permeability device behind a weak promoter | pSB3K3-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp-BBa_B0010-BBa_B0030-BBa_I746200-BBa_B0012 | 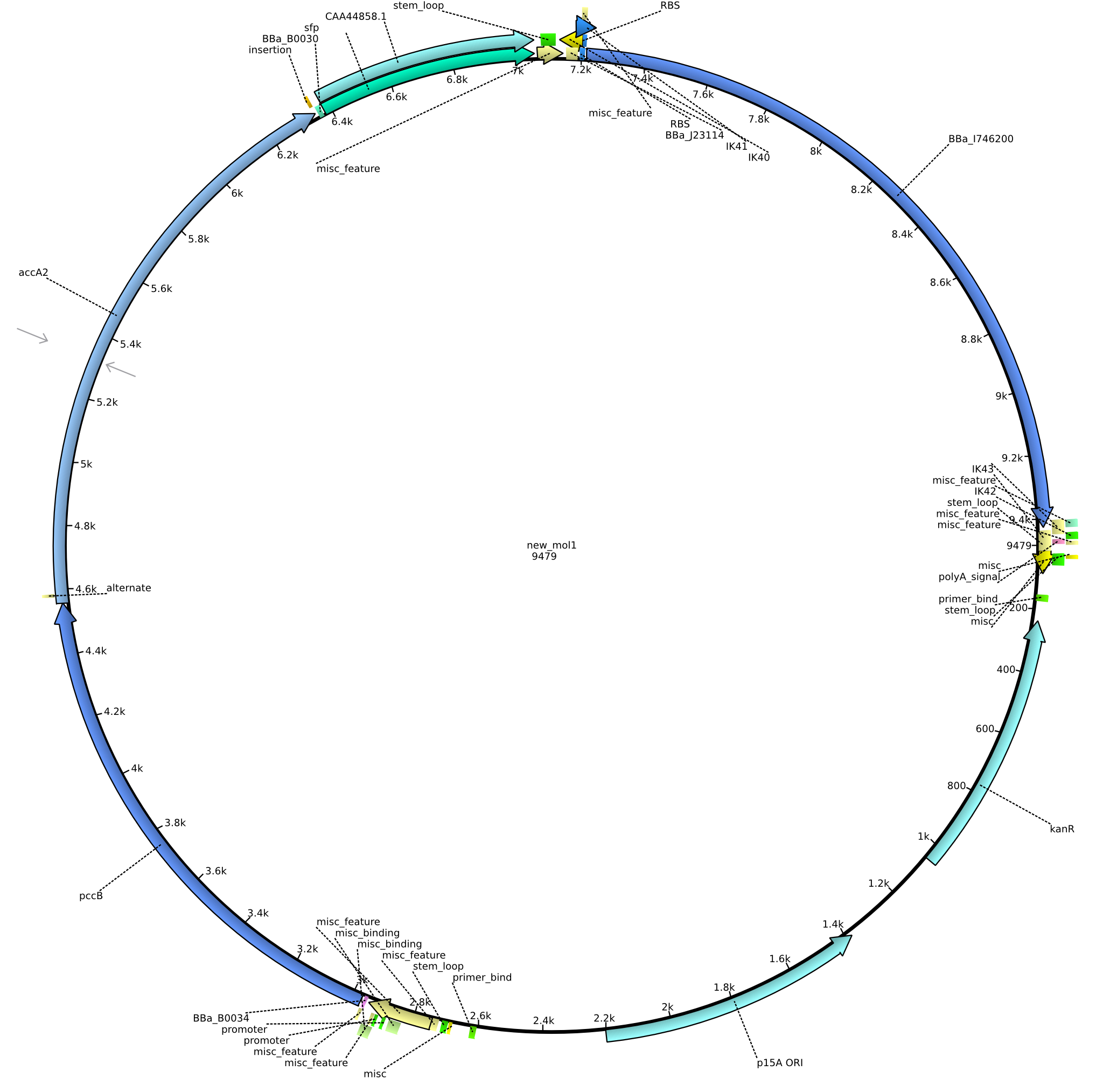 | pIK9 |
| pIK10 | 2013-09-18 | Methylmalonyl-CoA pathway with sfp and the permeability device behind a weak promoter, for submission | pSB1C3-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp-BBa_B0010-BBa_B0030-BBa_I746200-BBa_B0012 | 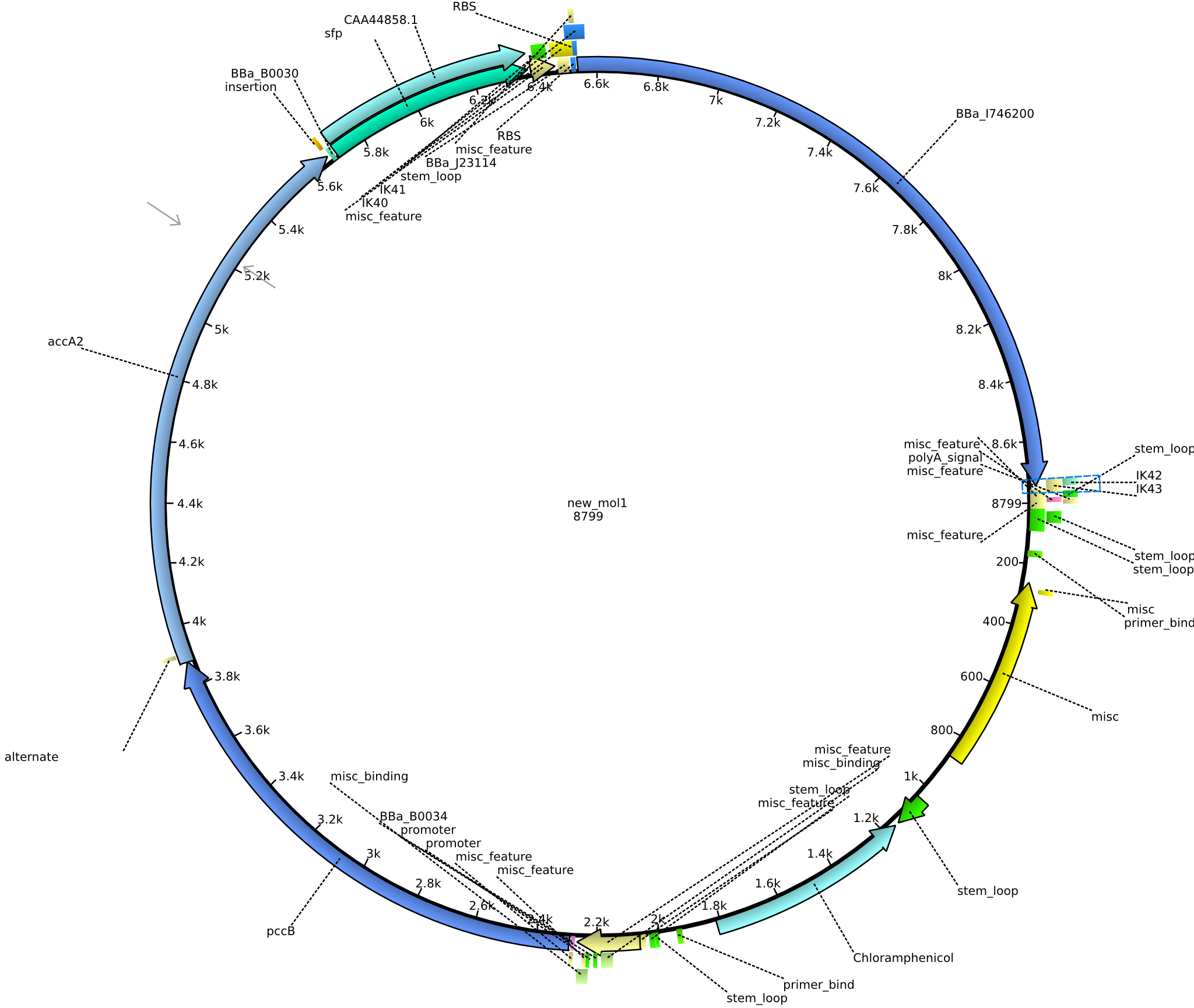 | pIK10 |
| pIK11 | 2013-09-18 | Methylmalonyl-CoA pathway with sfp, for submission | pSB1C3-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp | 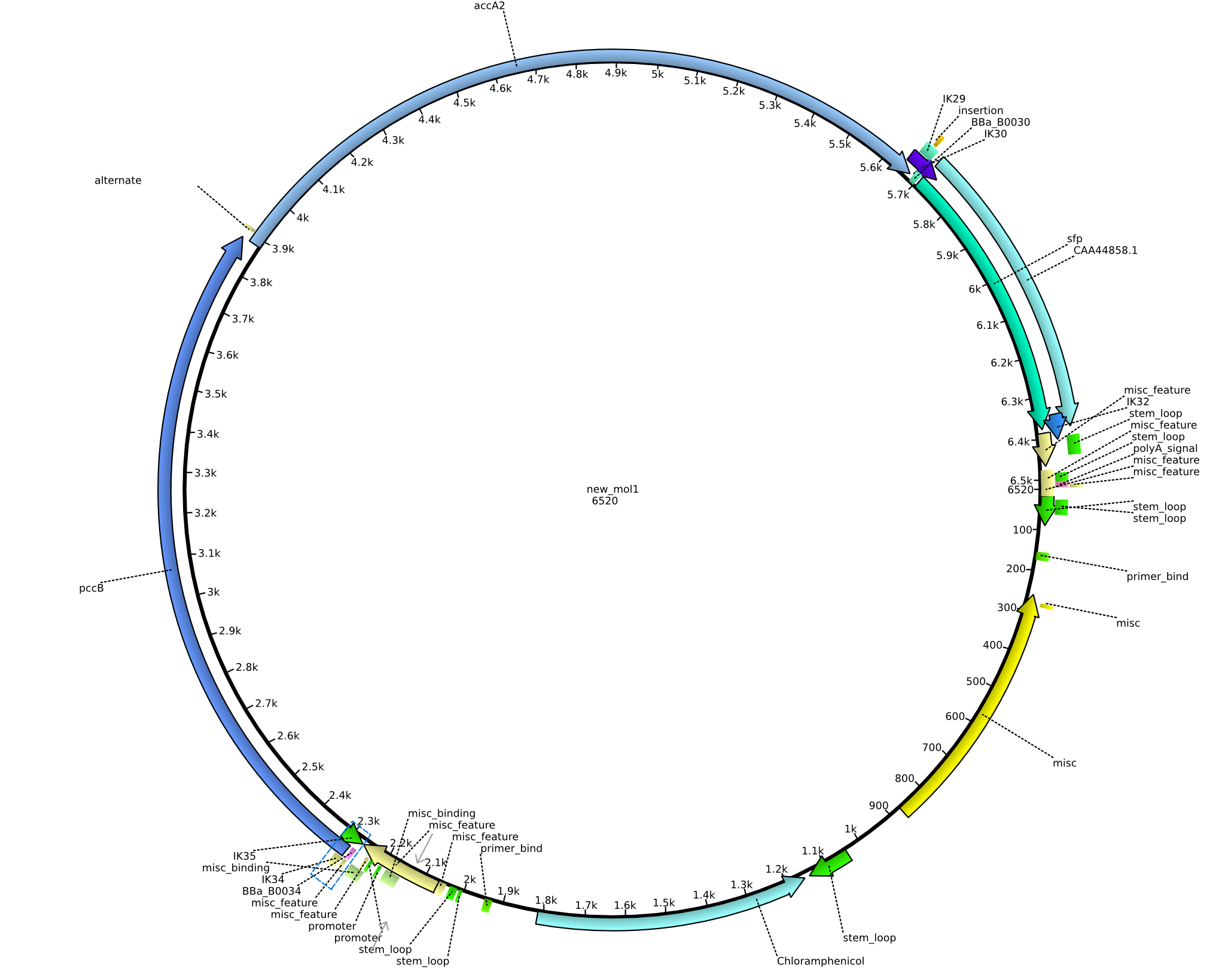 | pIK11 |
| pHM01 | 2013-04-29 | DelH construct assembled by Restriction Ligation Strategy | pSB6A1-BBa_K206000(AraC)-DelH-BBa_I732019(lacZ)-B0015(stop-codon) | 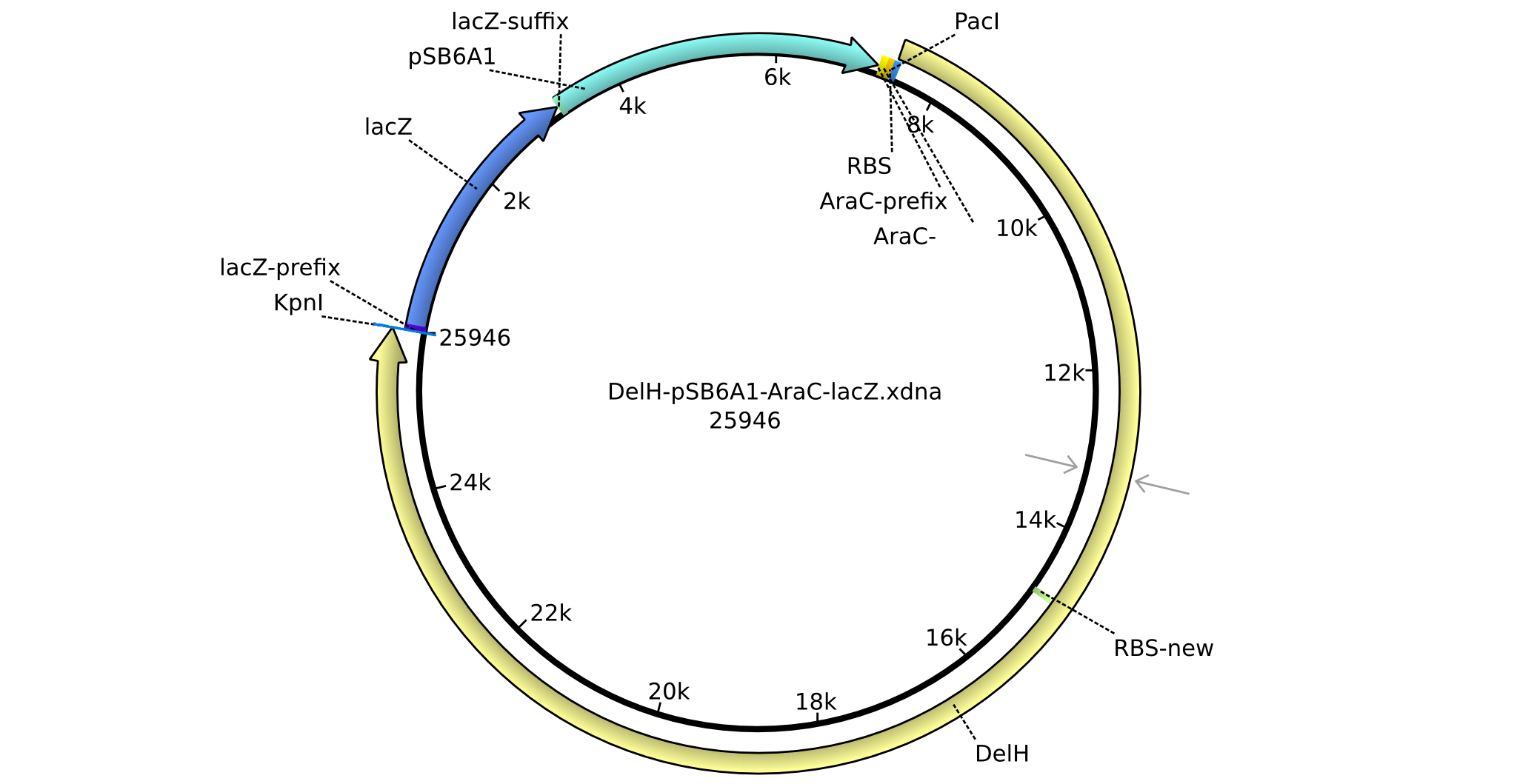 | pHM01 |
| pHM02 | 2013-07-08 | DelH construct assembled by Gibson Assemby | pSB6A1-R0010(lacI)-B0035(RBS)-DelH-B0034(RBS)-E1010(mRFP)-B0015(stop) | 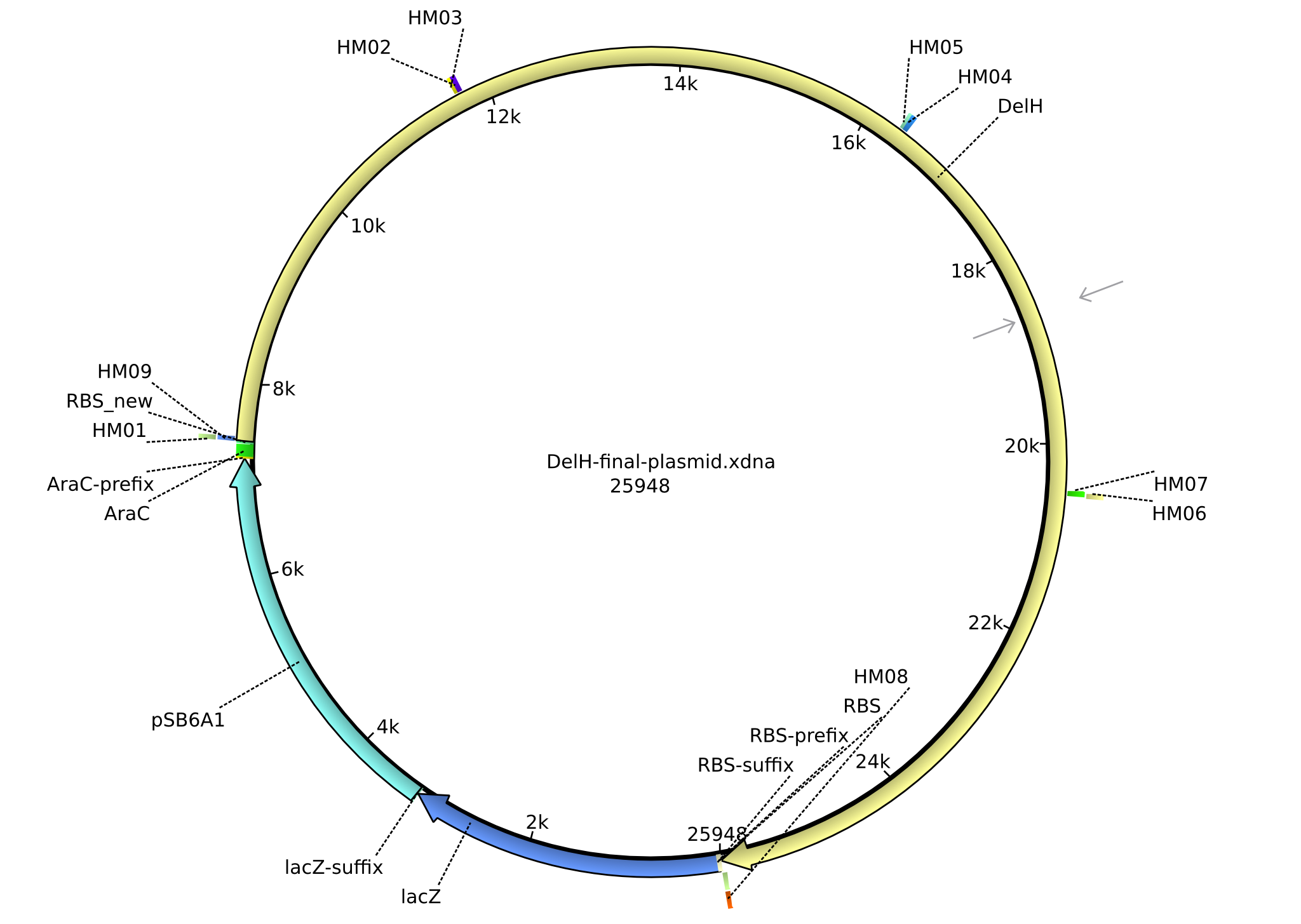 | pHM02 |
| pHM03 | 2013-04-29 | DelH construct assembled by Gibson Assemby for existing backbone | pSB6A1-R0010(lacI)-B0034(RBS)-DelH-B0035(RBS)-E1010(mRFP)-B0015(stop) | 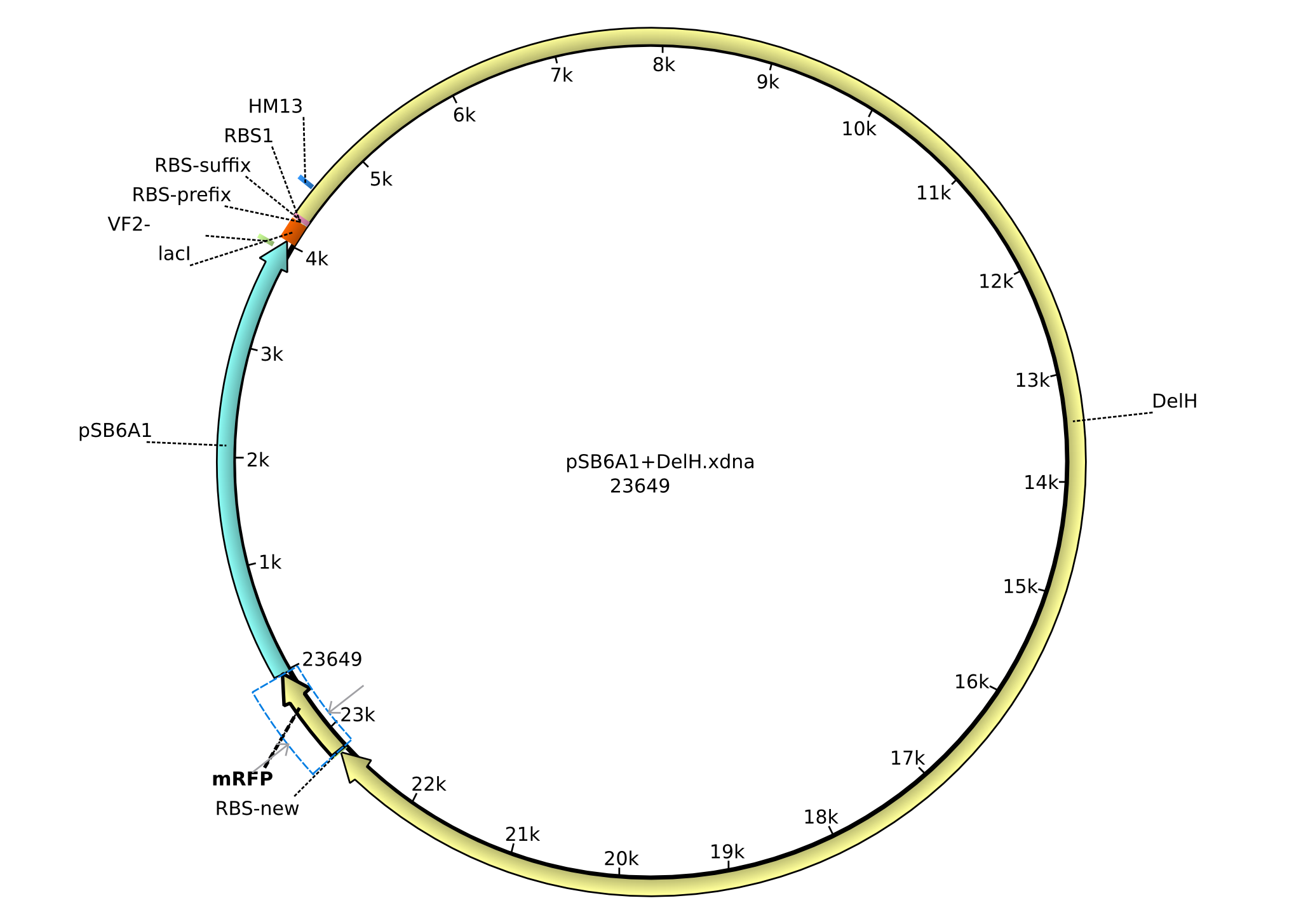 | pHM03 |
| pHM04 | 2013-08-12 | DelH construct assembled by Gibson Assemby without mRFP | pSB6A1-R0010(lacI)-B0034(RBS)-DelH-B0015(stop) | 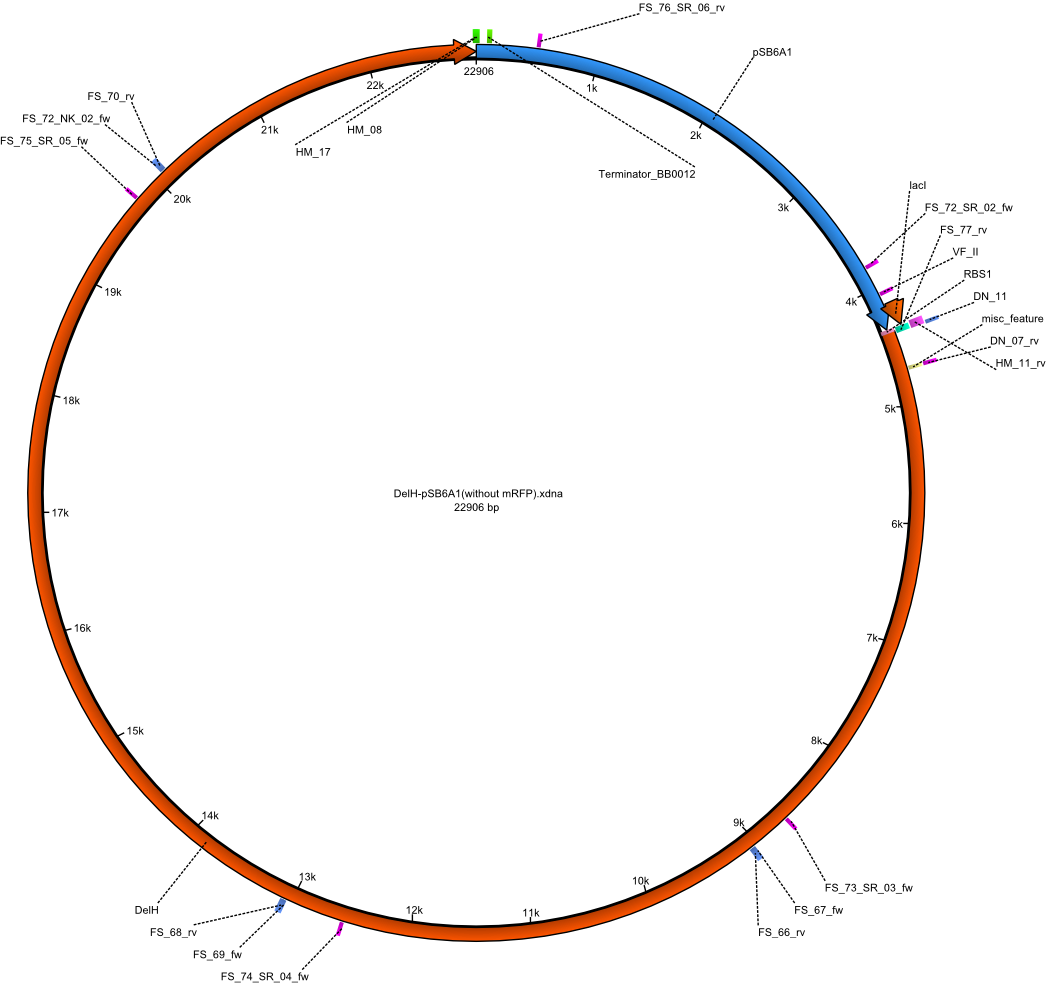 | pHM04 |
| pHM05 | 2013-08-12 | DelH construct assembled by Gibson Assemby with tetracycline resistance, without mRFP | pSB6A1-R0010(lacI)-DelH-B0015(stop)-pSB1T3(tetR) | 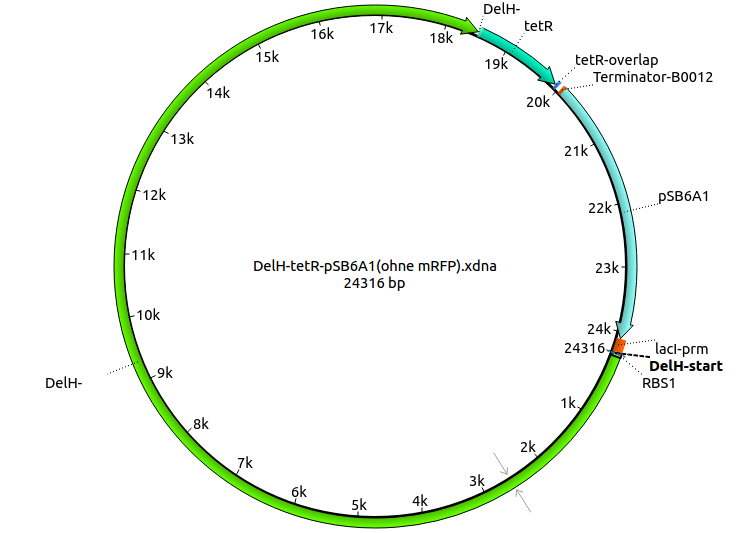 | pHM05 |
| pFSN | 2013-06-28 | DelRest, Delftibactin cluster from D. Acidovorans | pSB4K5-lacZ-BBa_B0035-DelA-DelB-DelC-DelE-DelF-DelG-DelO-DelP-DelL-BBa_B0034 | 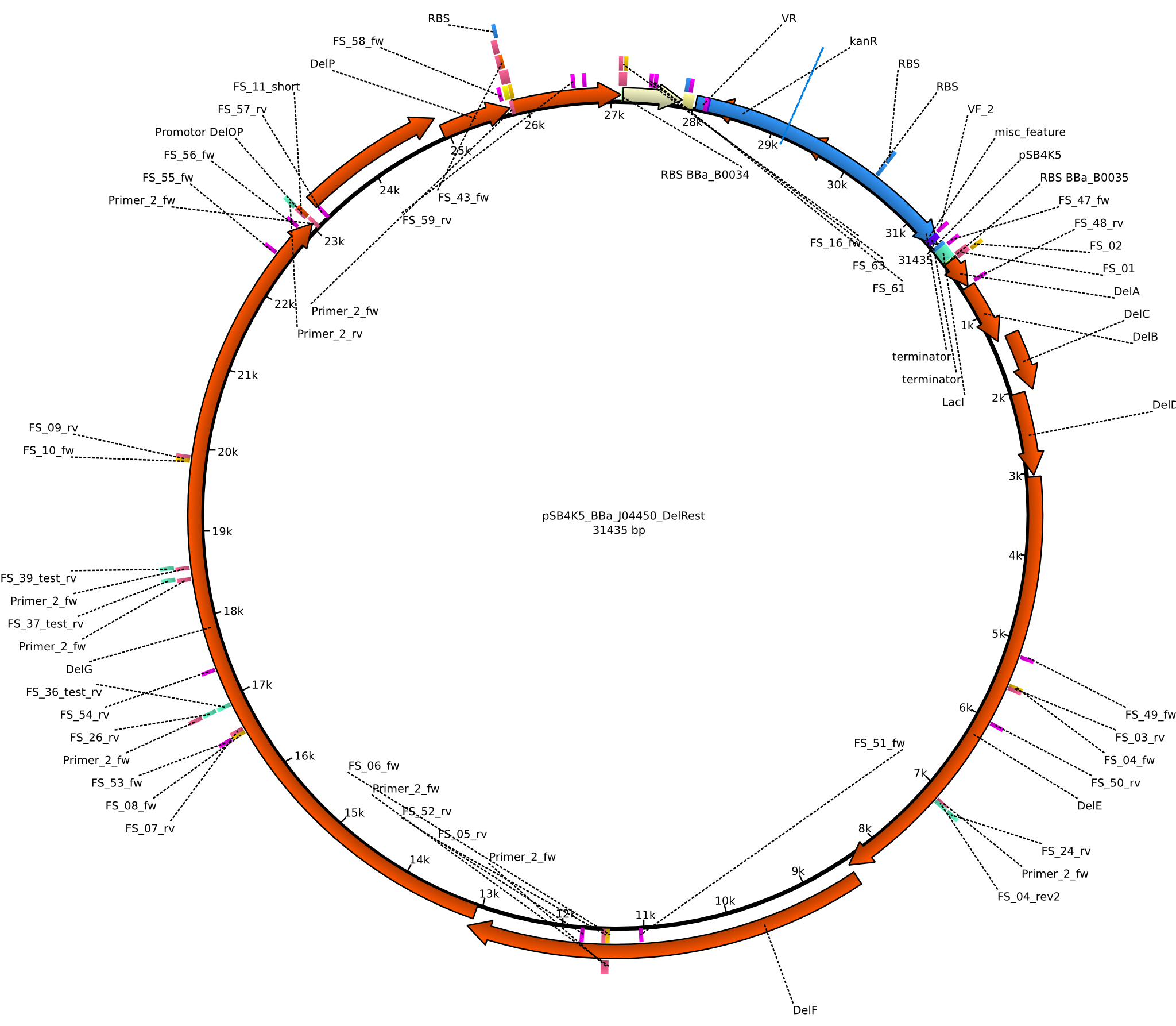 | pFSN |
| pFS_02 | 2013-09-26 | DelH construct assembled by Gibson Assemby with weak Promotor, weak RBS | pSB6A1-BBa_J23114-BBa_B0032-DelH | 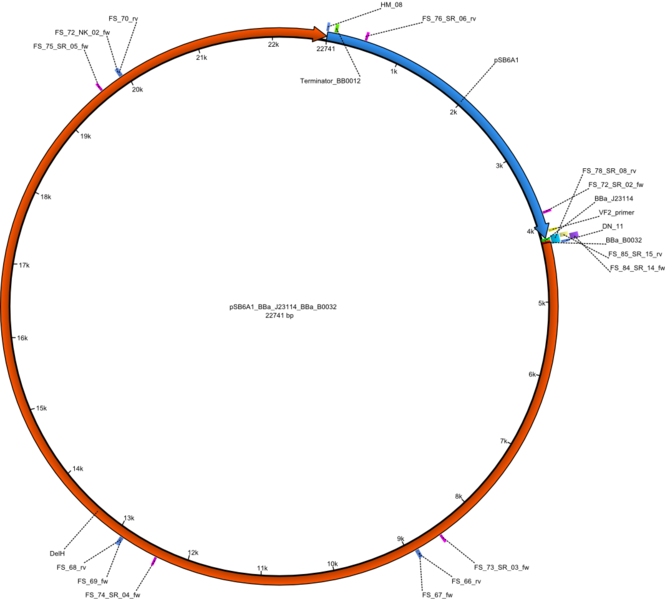 | pFS_02 |
| pFS_03 | 2013-09-26 | Helper plasmid for DelH restriction cloning, DelH, flanked by KpnI and BamHI site, without promotor, | pSB4K5-DelH | 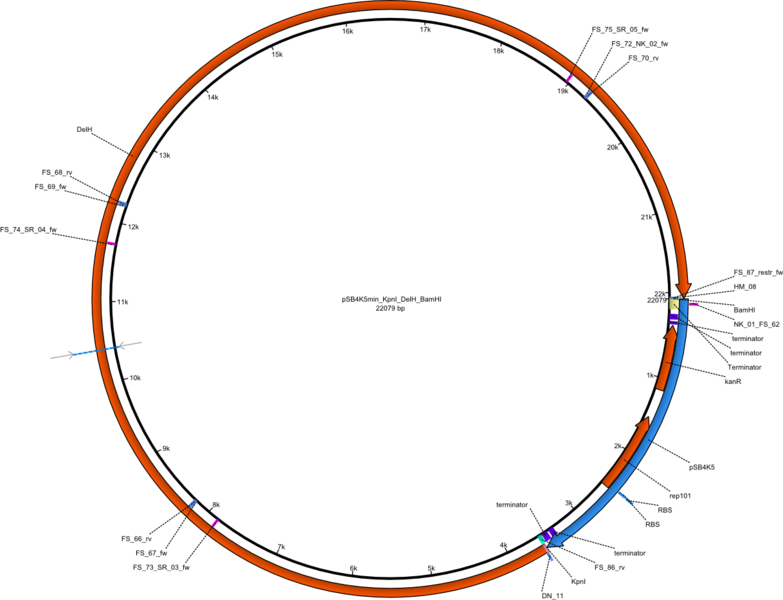 | pFS_03 |
| pFS_04 | 2013-09-26 | Target plasmid for DelH restriction cloning, ccdB cassette, flanked by KpnI and BamHI site | pSB6A1-BBa_J23114-BBa_B0032-ccdBcassette |  | pFS_04 |
| pFS_05 | 2013-09-26 | DelH construct assembled by restriction cloning using pFS_03 and pFS-04 DelH, flanked by KpnI and BamHI site, under weak promotor, | pSB6A1-BBa_J23114-BBa_B0032-DelH |  | pFS_05 |
Module Shuffling
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
| pIK03 | 2013-mm-dd | NRPS for Pro-Leu | pSB1C3+TycB1dComdC+TycC6 | 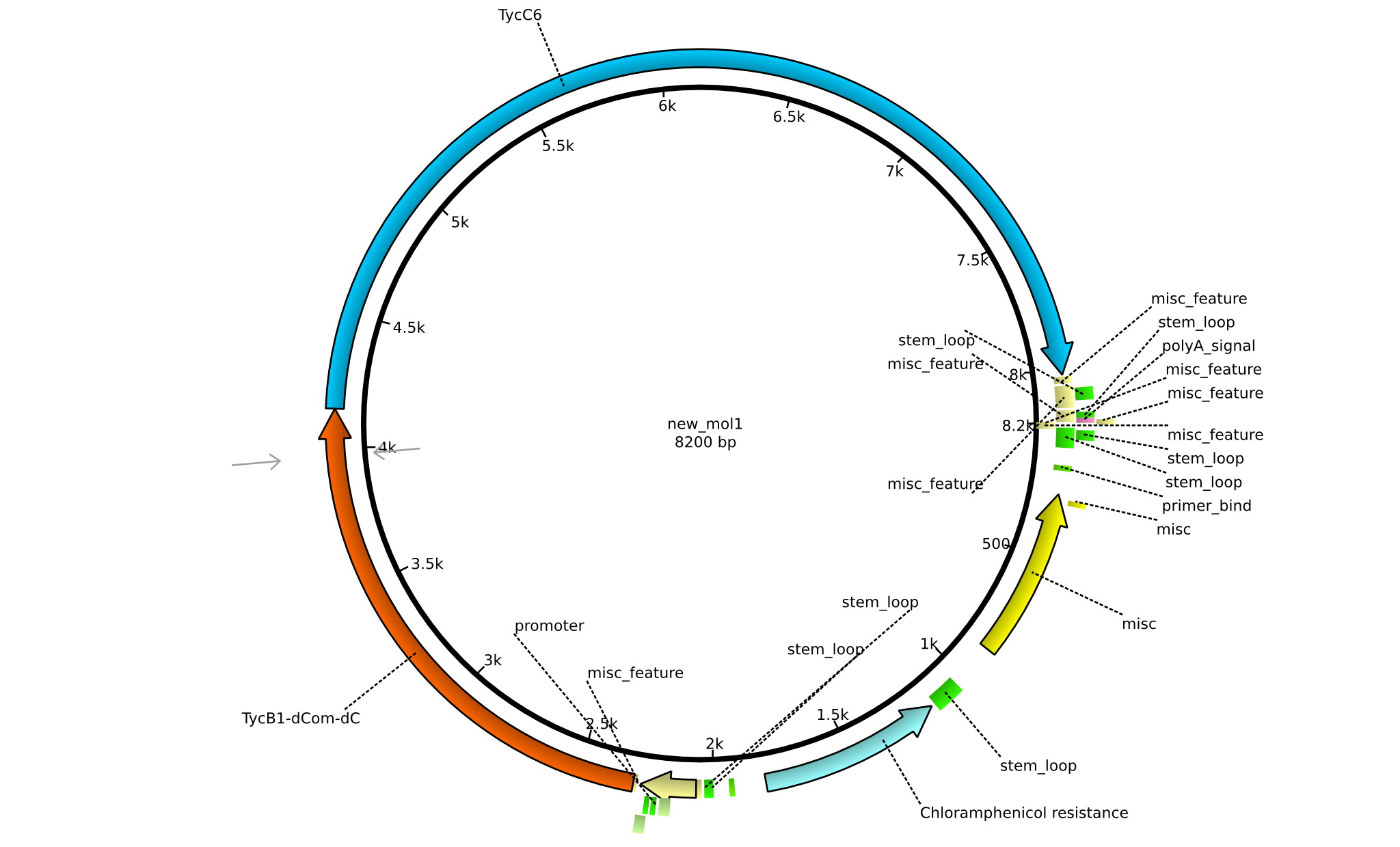 | pIK03 |
| pIK04 | 2013-mm-dd | NRPS for Phe-Orn-Leu | pSB1C3+TycAdCom+TycC5-TycC6 | 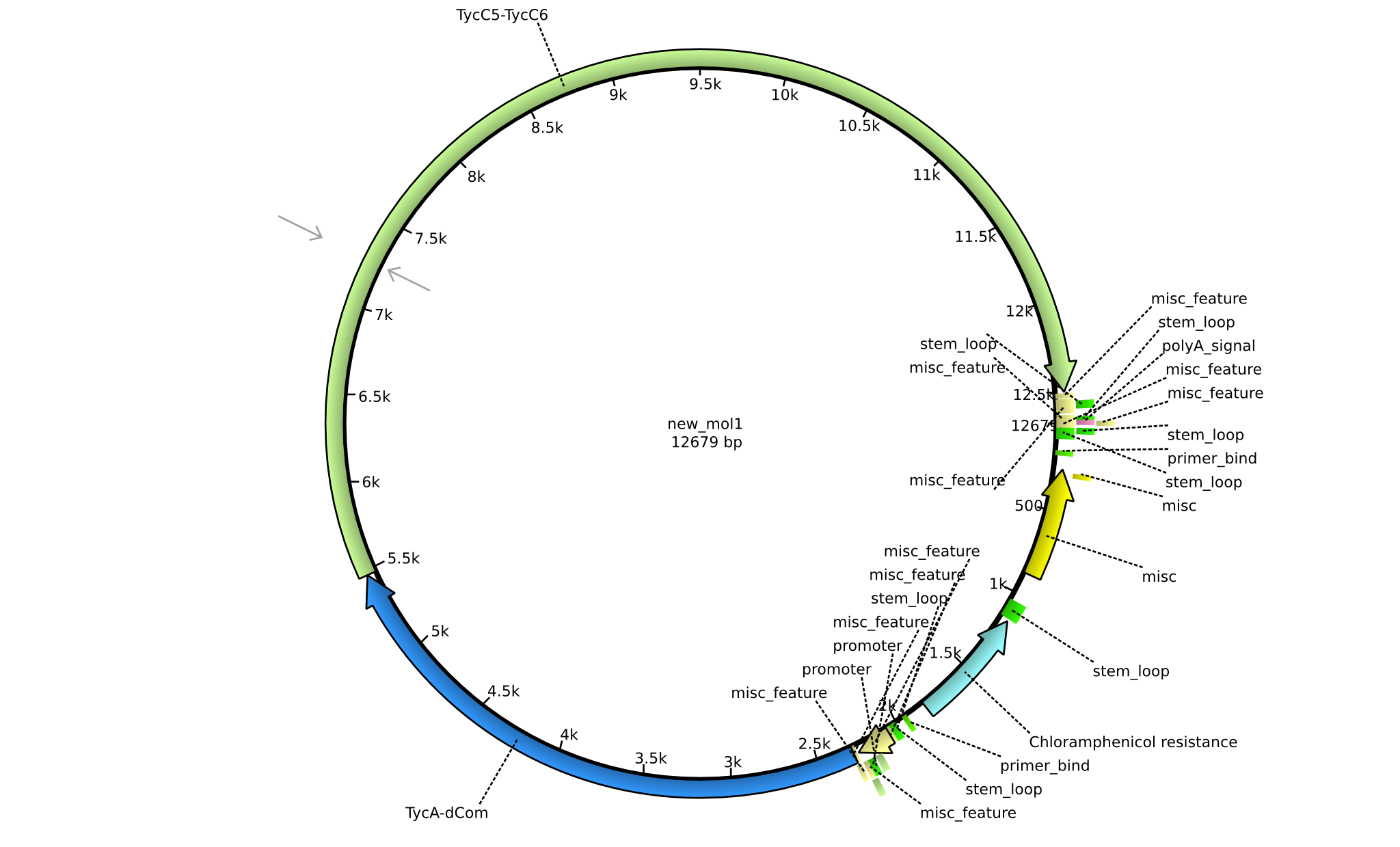 | pIK04 |
| pIK05 | 2013-mm-dd | NRPS for Pro-Orn-Leu | pSB1C3+TycB1dComdC+TycC5-TycC6 | 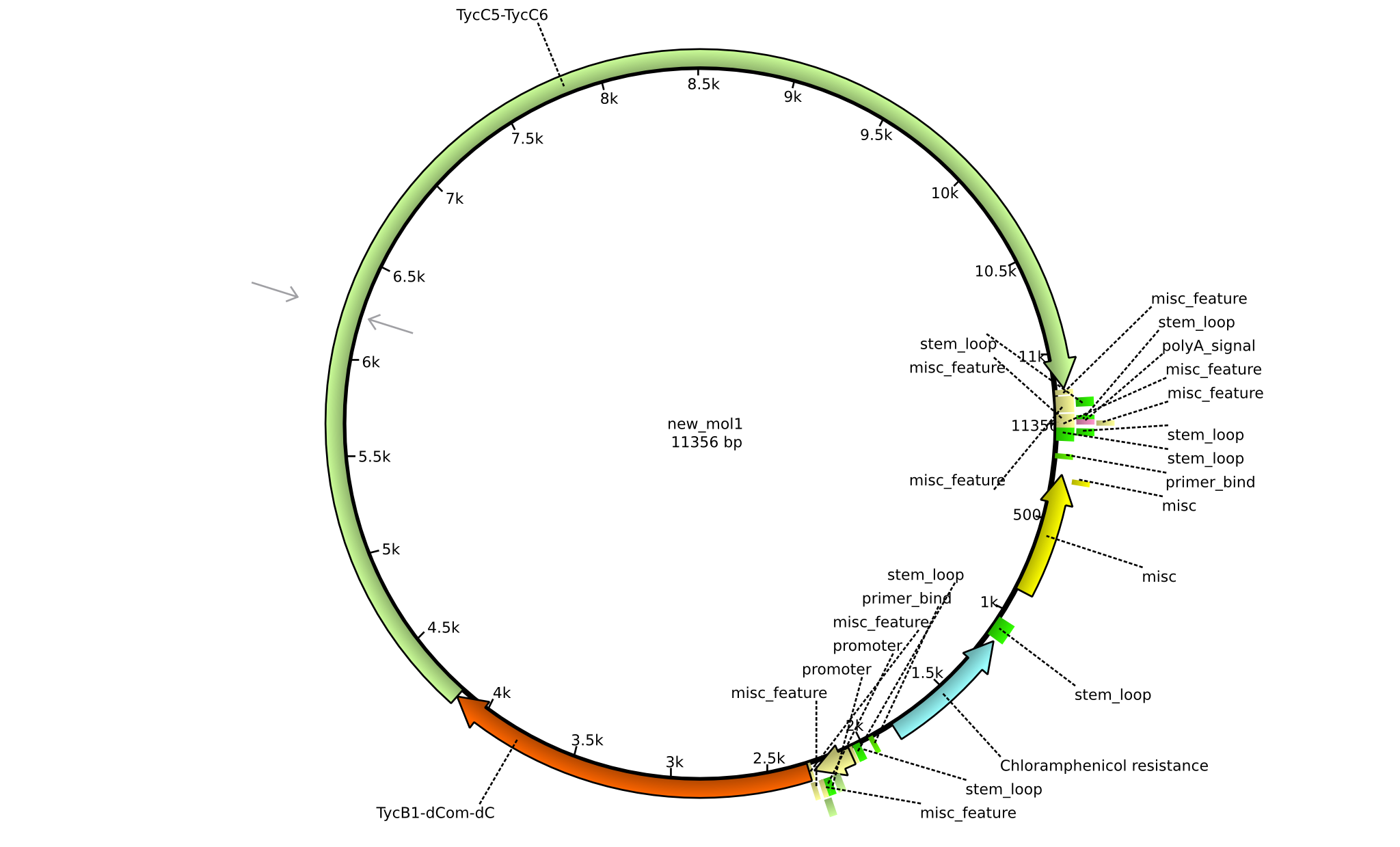 | pIK05 |
| pPW01 | 2013-mm-dd | NRPS for Orn-Pro-Phe-Leu | pSB1C3+TycC5+TycB1dCom-C(TycB2)+TycAdCom+TycC6 | 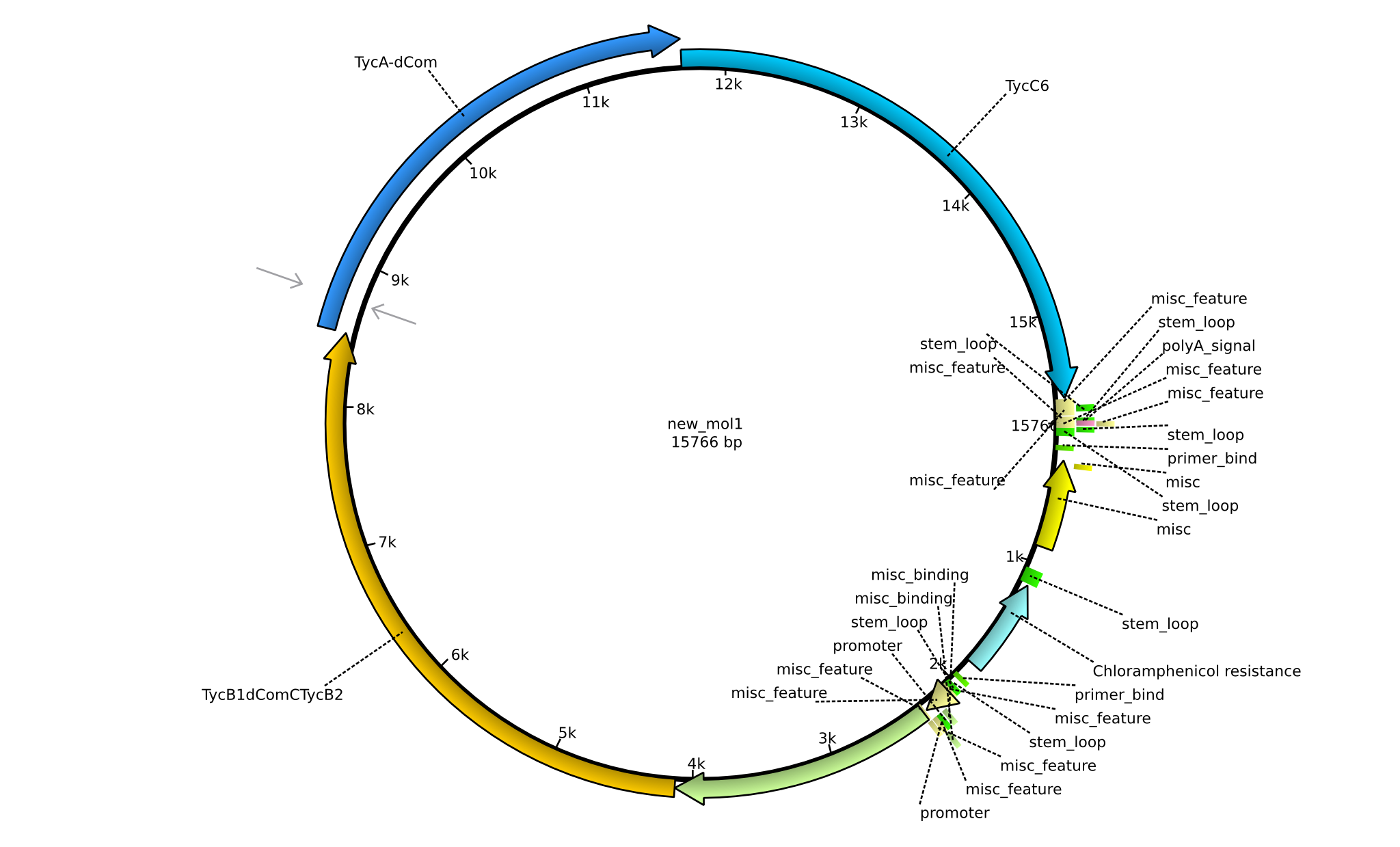 | pPW01 |
| pPW02 | 2013-mm-dd | NRPS for Orn-Pro-Phe-Leu | pSB1C3+TycC5+TycB1dCom-C(TycB2)+TycAdE+TycC6 | 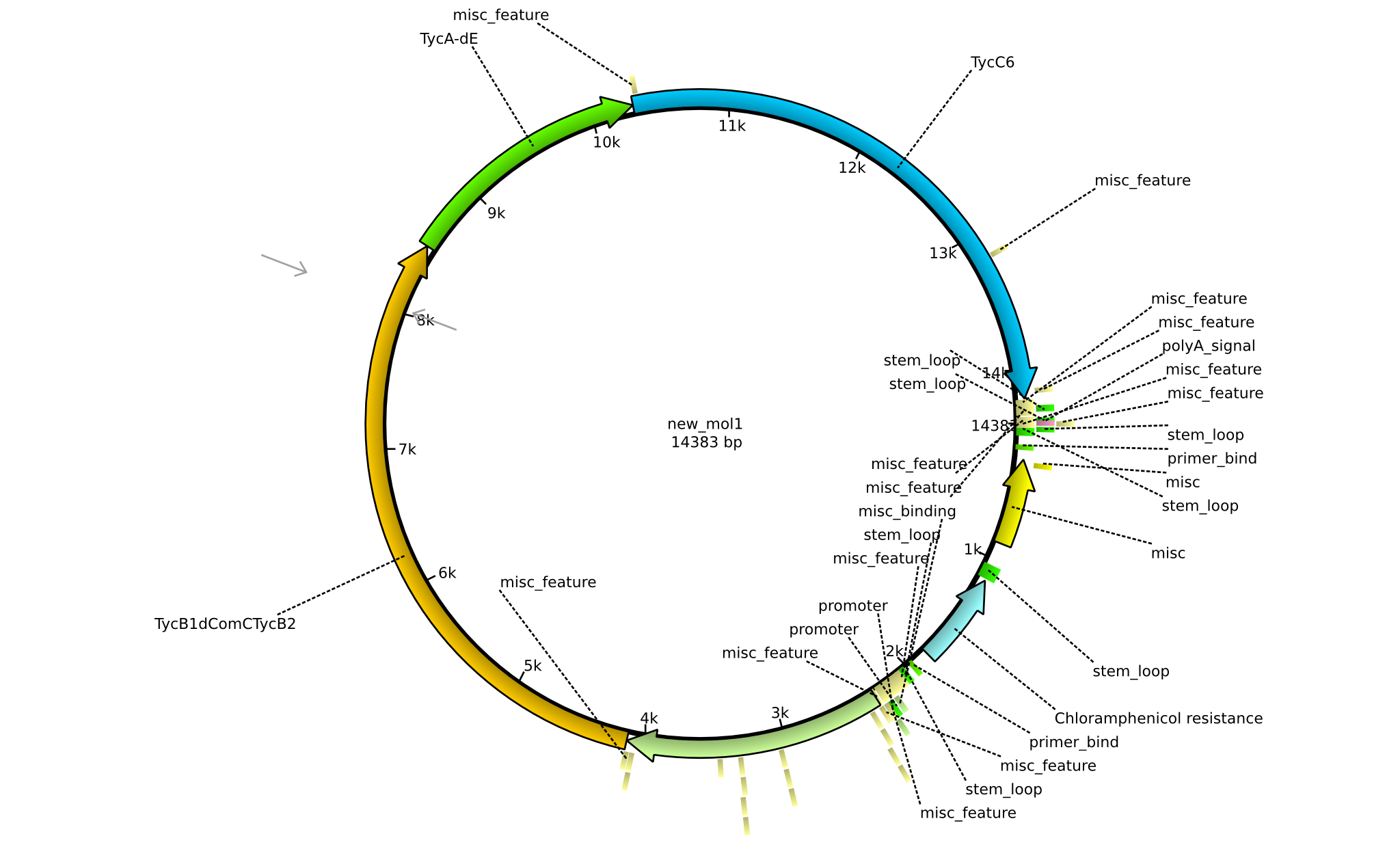 | pPW02 |
| pPW03 | 2013-08-18 | NRPS for Phe-Indigoidine | pSB1C3+TycA+C(TycC2)+IndC | 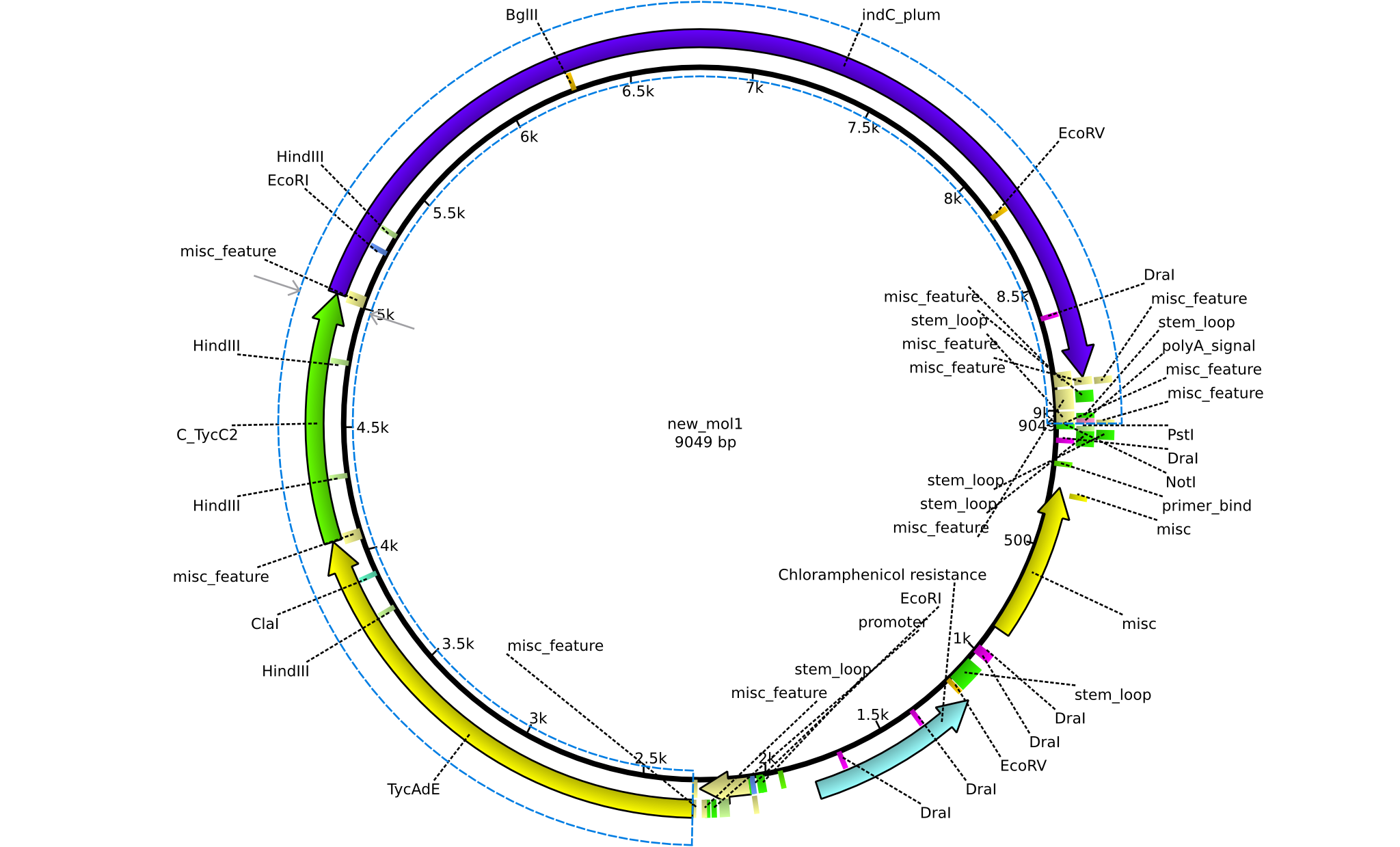 | pPW03 |
| pPW04 | 2013-08-18 | NRPS for Asn-Indigoidine | pSB1C3+TycC1-C(TycC2)+IndC | 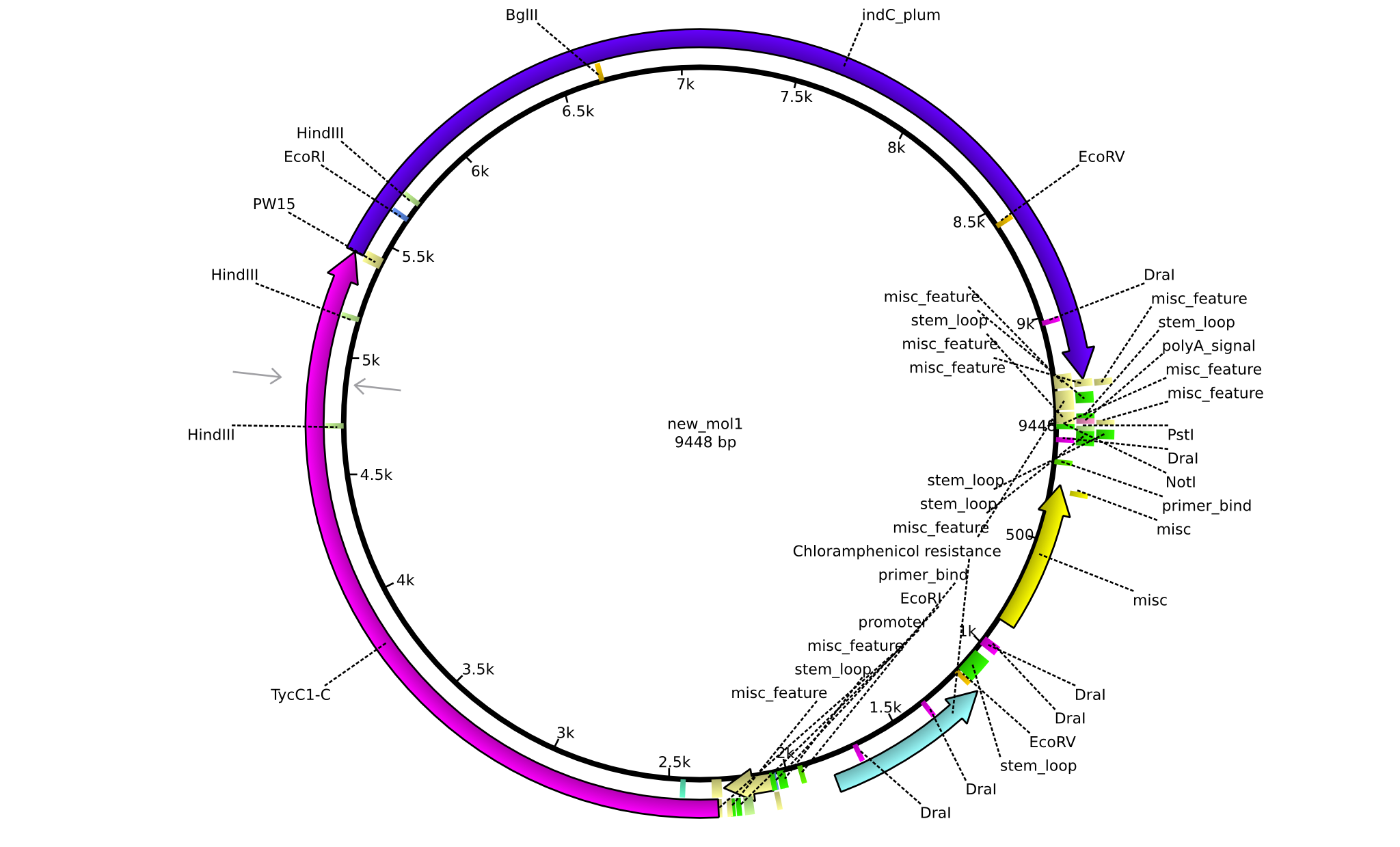 | pPW04 |
| pPW05 | 2013-08-18 | NRPS for Val-Indigoidine | pSB1C3+TycC4+C(TycC2)+IndC | 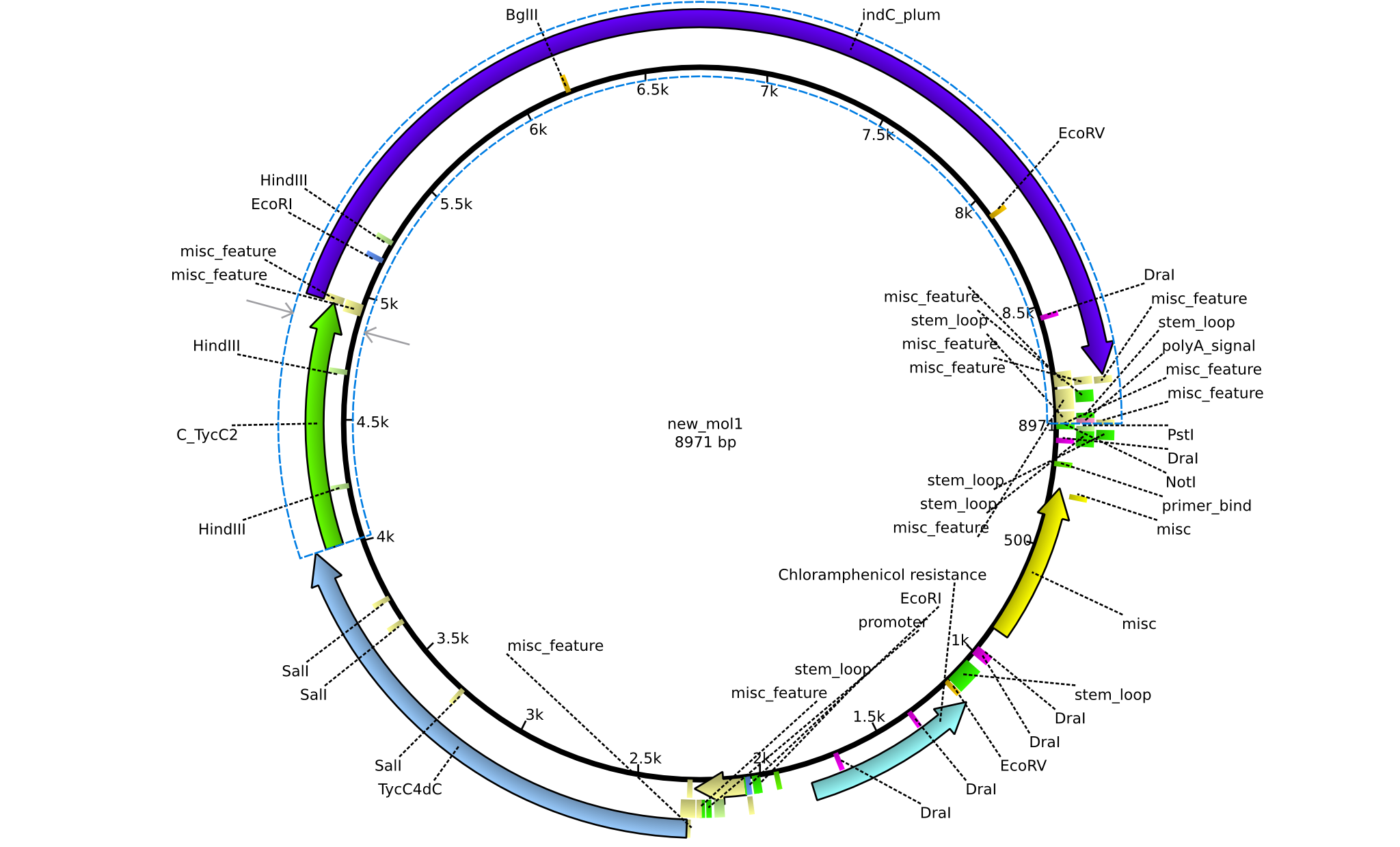 | pPW05 |
| pPW06 | 2013-09-07 | NRPS for Orn-Val-Indigoidine | pSB1C3+TycC5dC+C(TycC4)+TycC4dC+C(TycC2)+indC | 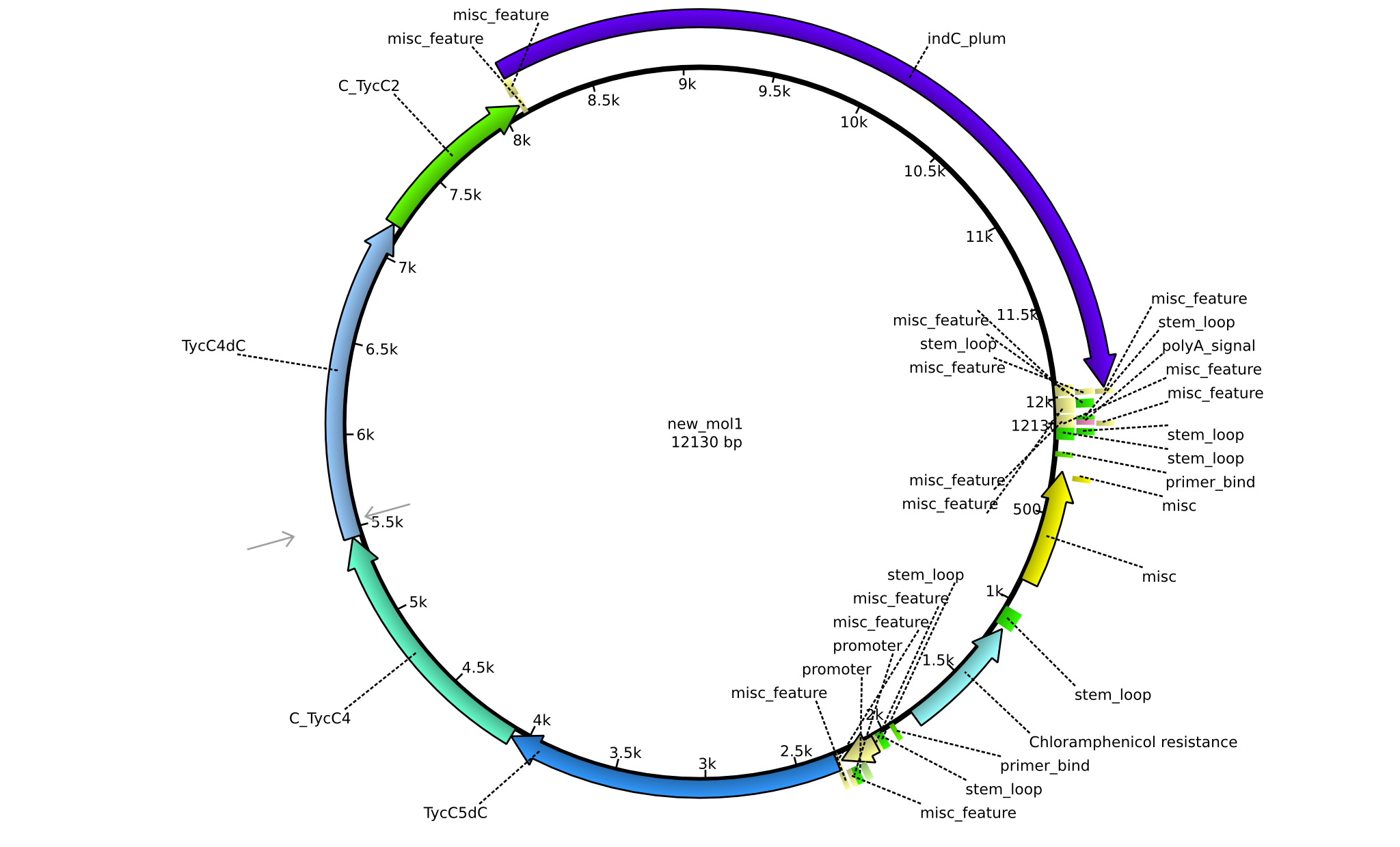 | pPW06 |
| pPW07 | 2013-09-07 | NRPS for Orn-Val-Val-Indigoidine | pSB1C3+TycC5dC+TycC4+C(TycC4)+TycC4dC+C(TycC2)+indC | 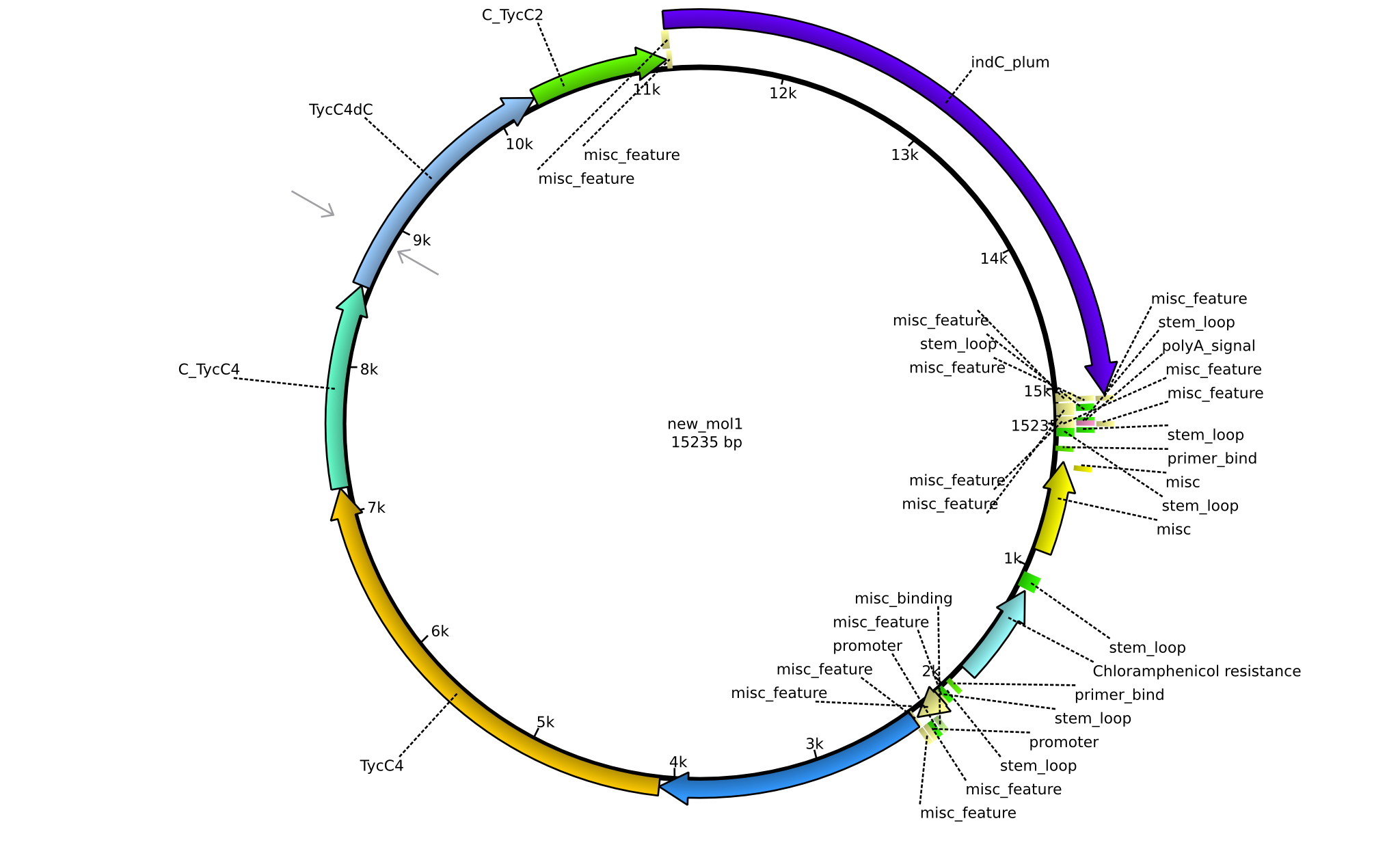 | pPW07 |
| pPW08 | 2013-09-07 | NRPS for Val-Val-Indigoidine | pSB1C3+TycC4dC+C(TycC4)+TycC4dC+C(TycC2)+indC | 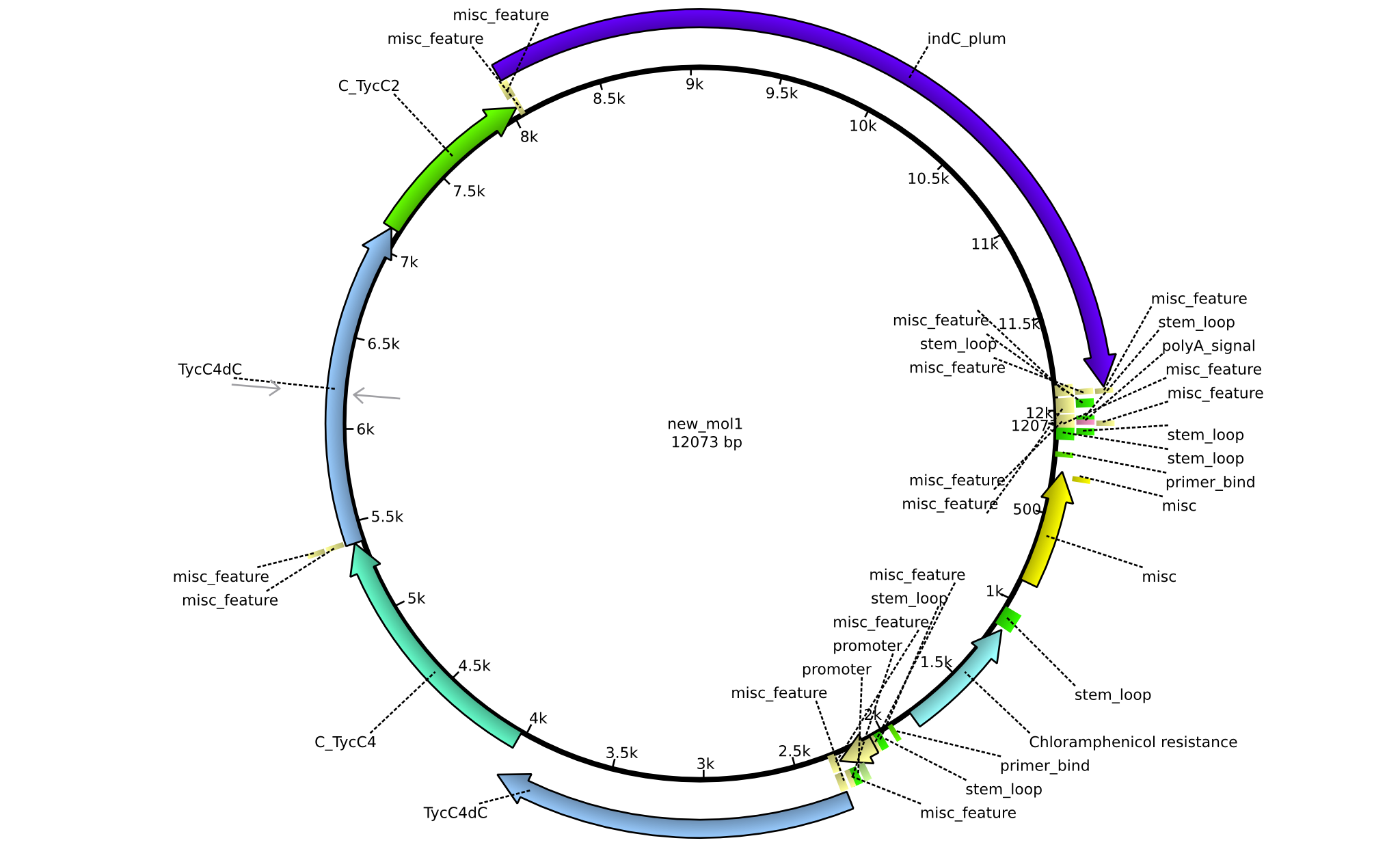 | pPW08 |
| pPW09 | 2013-09-07 | NRPS for Pro-Leu-Indigoidine | pSB1C3+TycB1dComdC+TycC6dTE+C(TycC2)+indC | 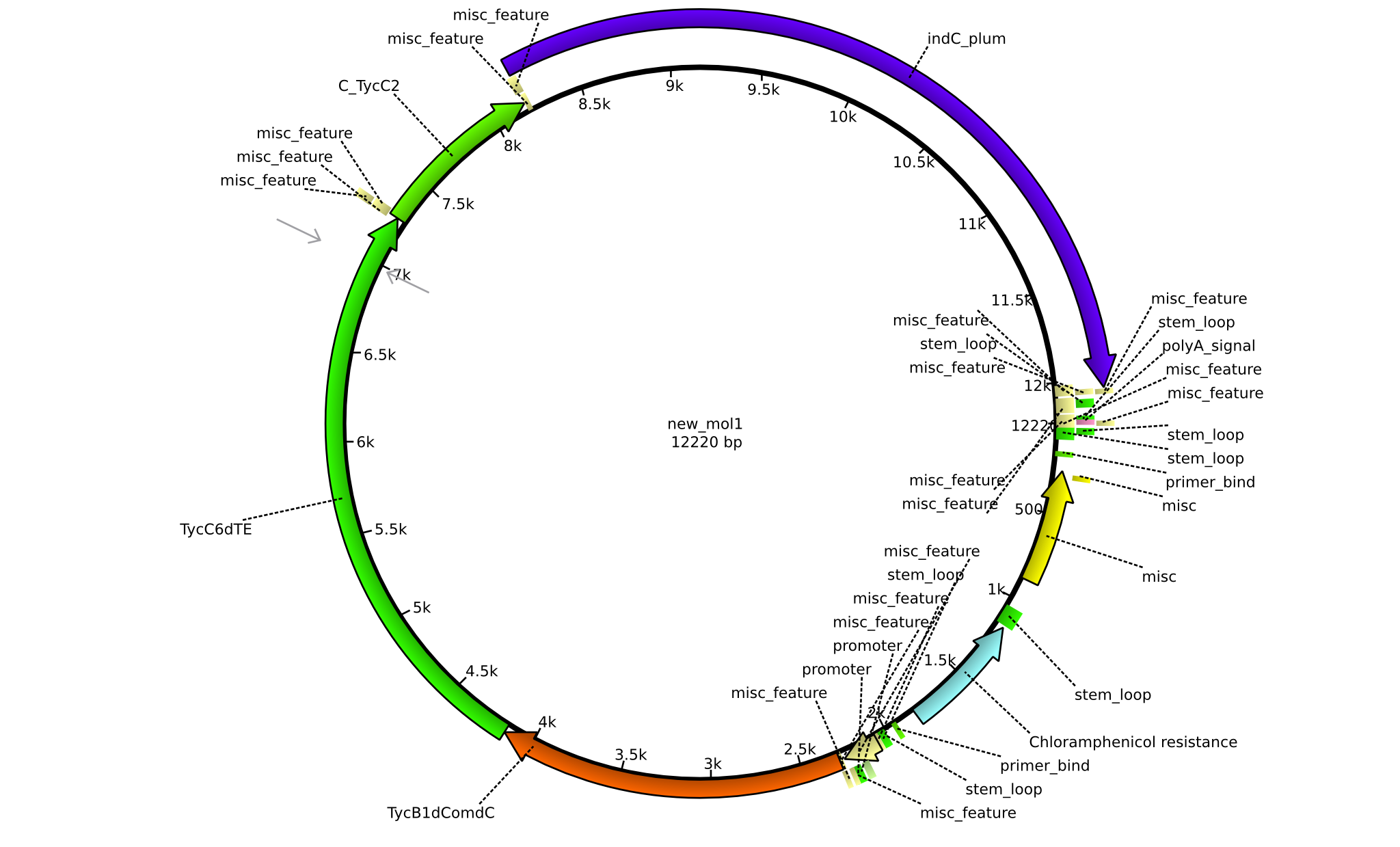 | pPW09 |
| pPW10 | 2013-09-07 | NRPS for Pro-Leu-Val-Indigoidine | pSB1C3+TycB1dComdC+TycC6dTE+C(TycC4)+TycC4dC+C(TycC2)+indC | 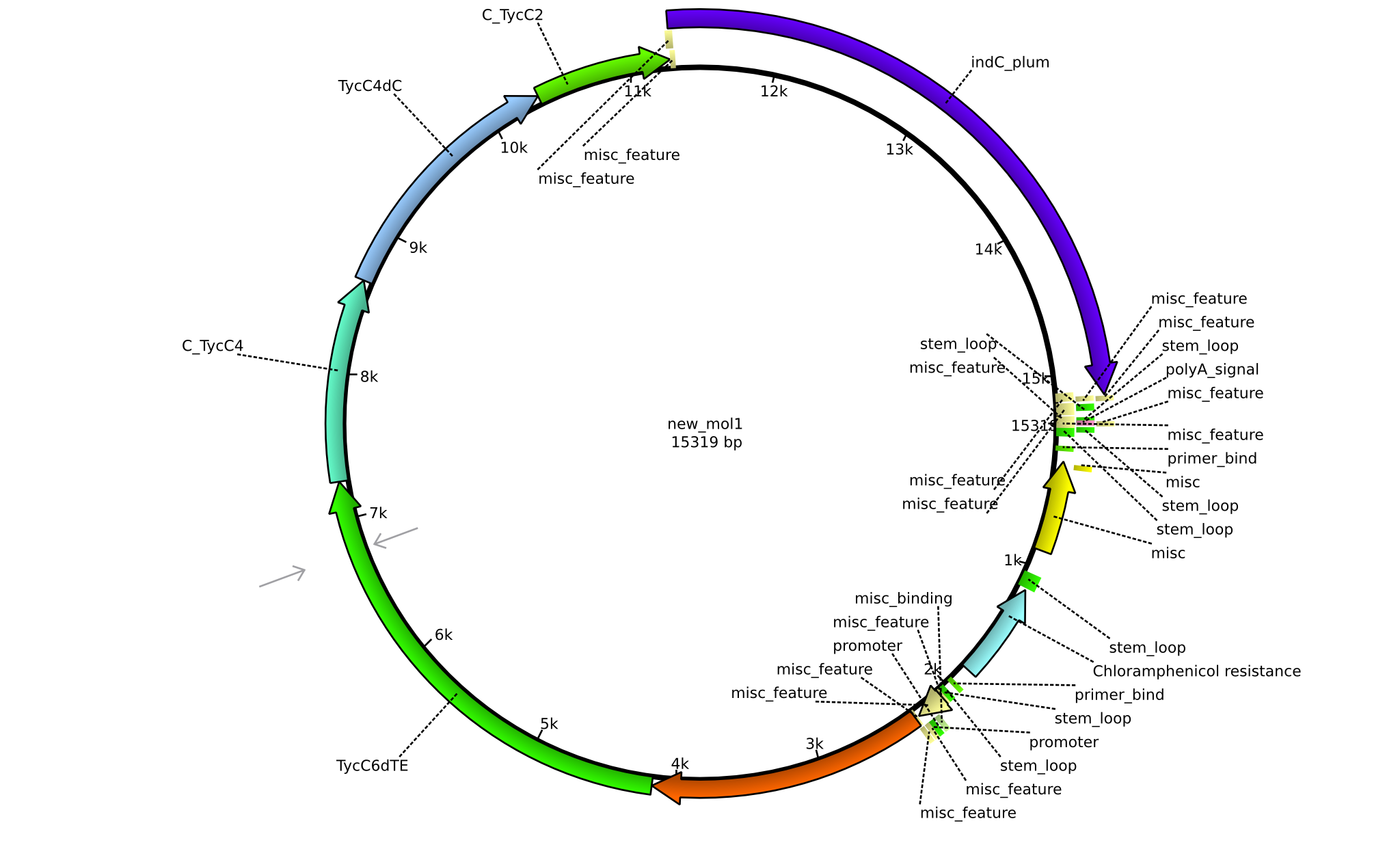 | pPW10 |
| pPW11 | 2013-09-07 | NRPS for Phe-Orn-Leu-Indigoidine | pSB1C3+TycAdCom+TycC5-TycC6dTE+C(TycC2)+indC | 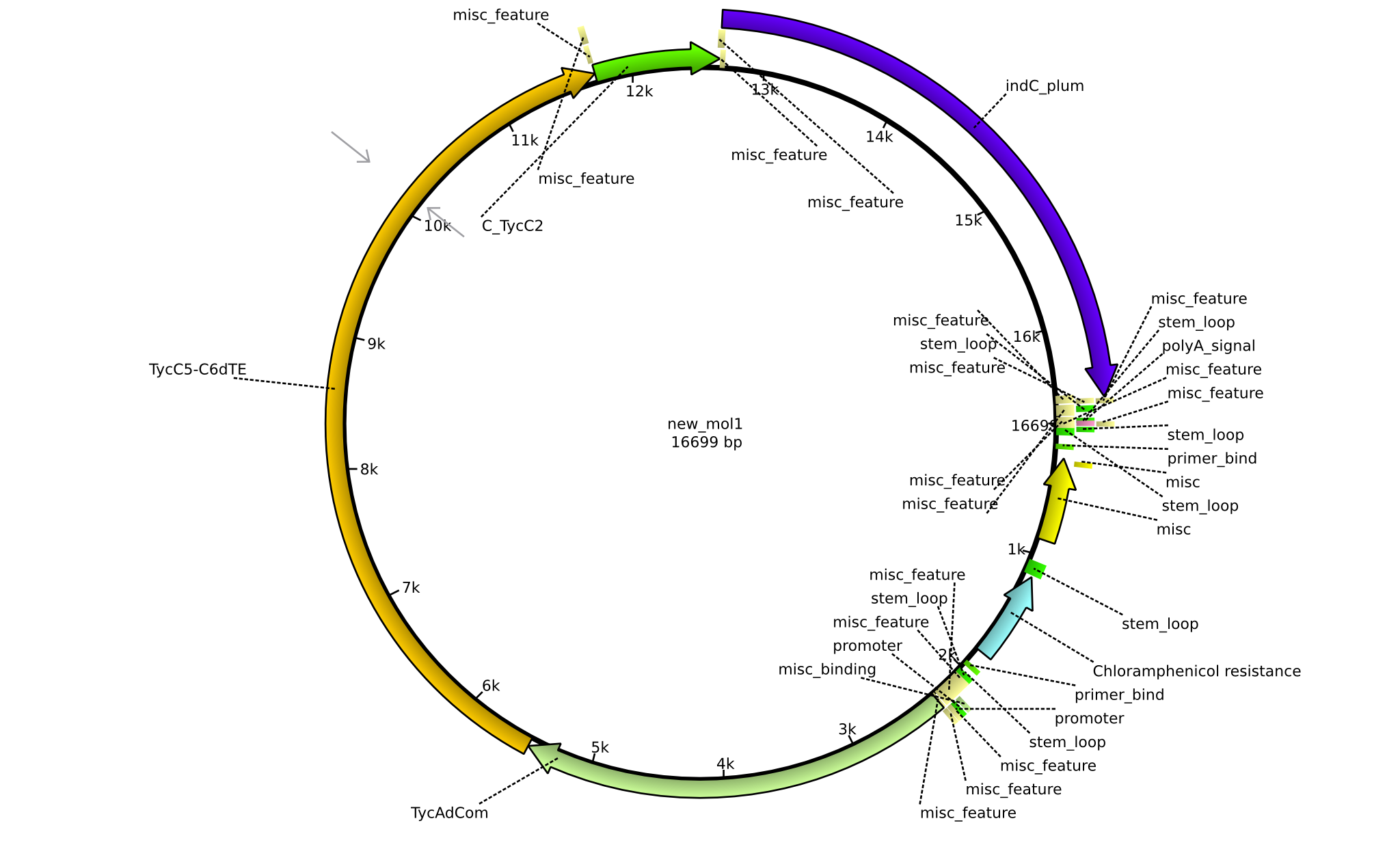 | pPW11 |
| pPW12 | 2013-09-07 | NRPS for Phe-Orn-Leu-Val-Indigoidine | pSB1C3+TycAdCom+TycC5-TycC6dTE+C(TycC4)+TycC4dC+C(TycC2)+indC | 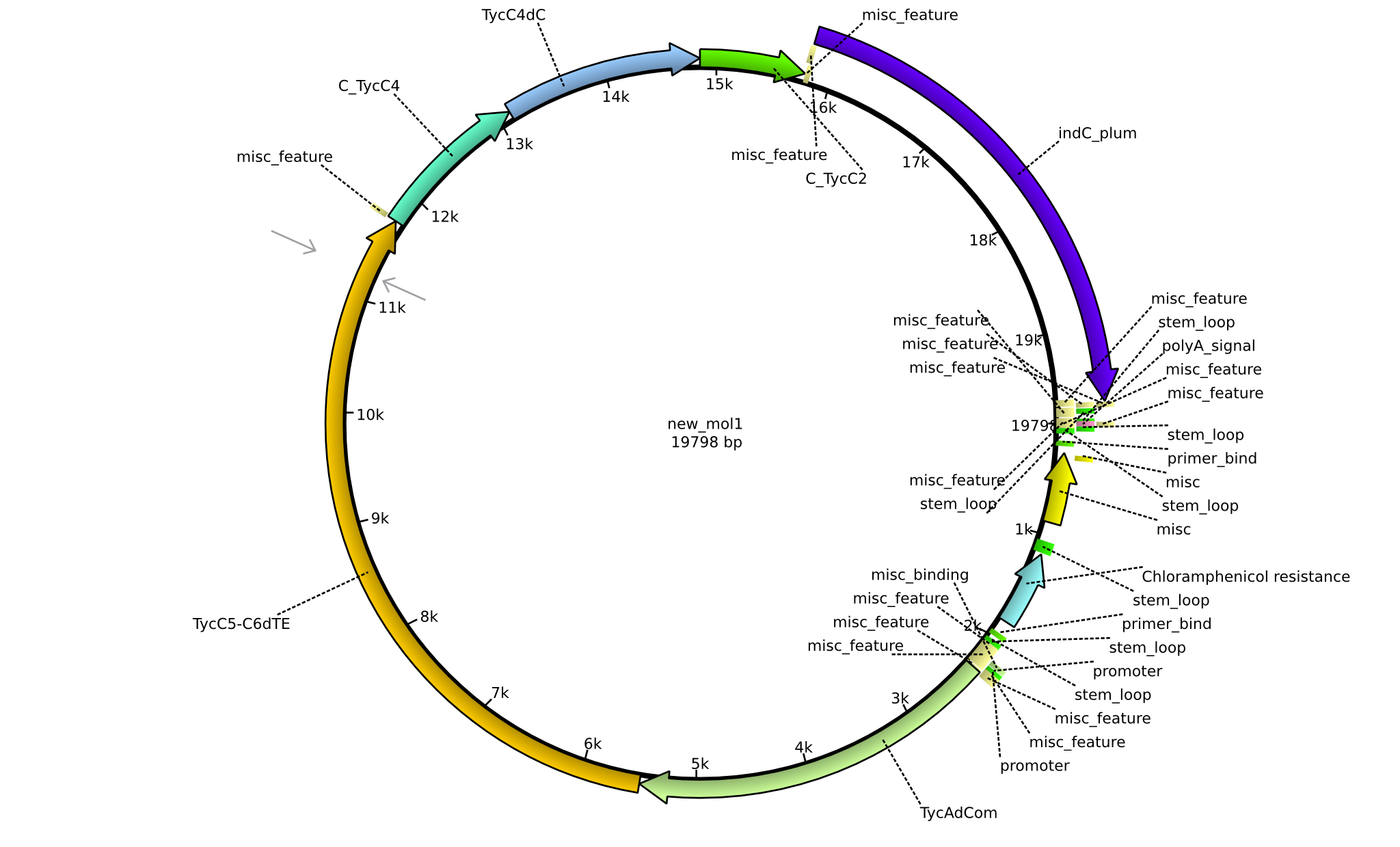 | pPW12 |
| pJS01 | 2013-09-18 | Helper plasmid for the tagging of NRPs with Indigoidine | pSB1C3+ccdB+C(TycC2)+indC | 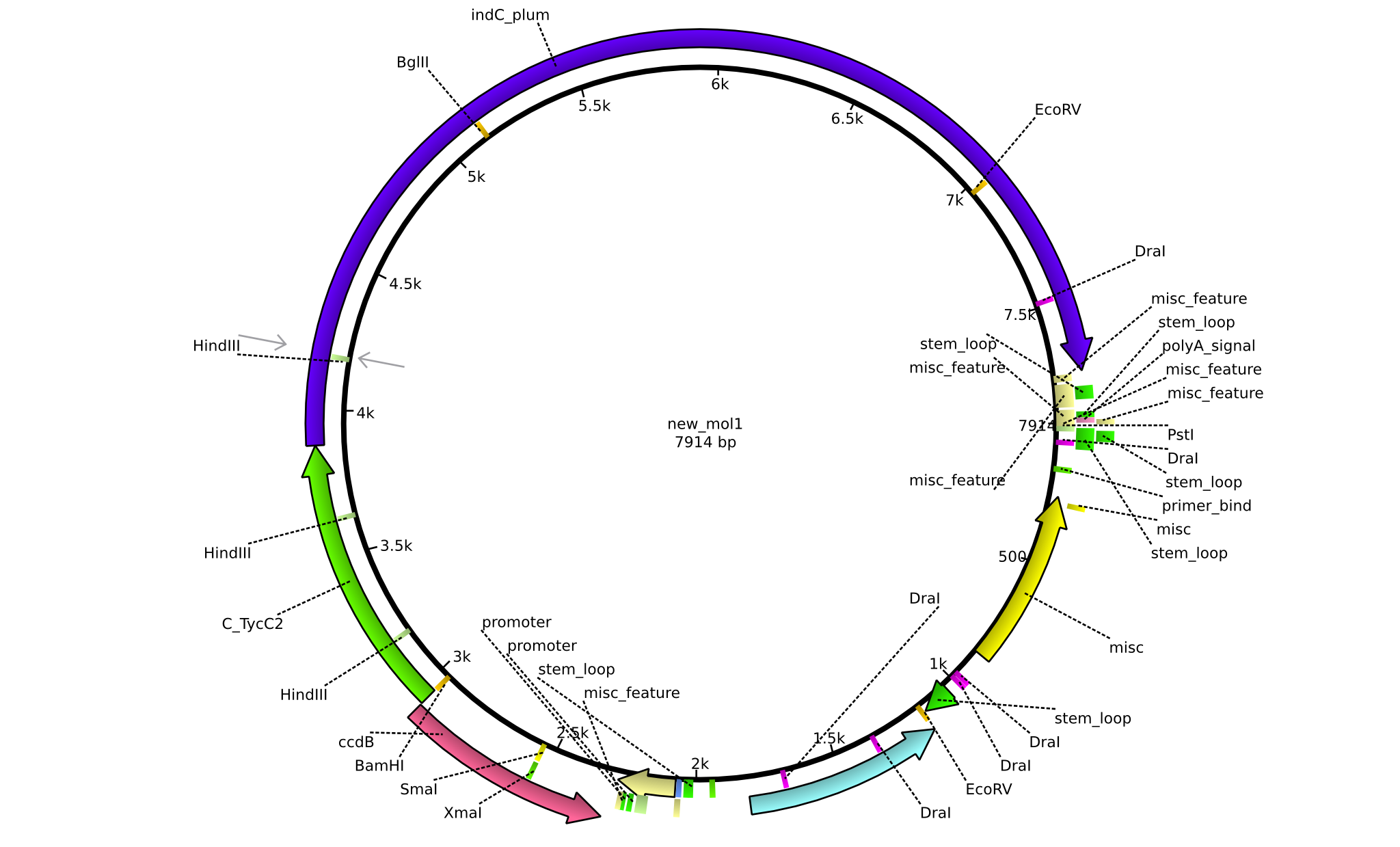 | pJS01 |
Linker variation
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
| pAR01 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with double short linkers | pSB1C3-TycC4dCom_s+TycC2_s+indC | 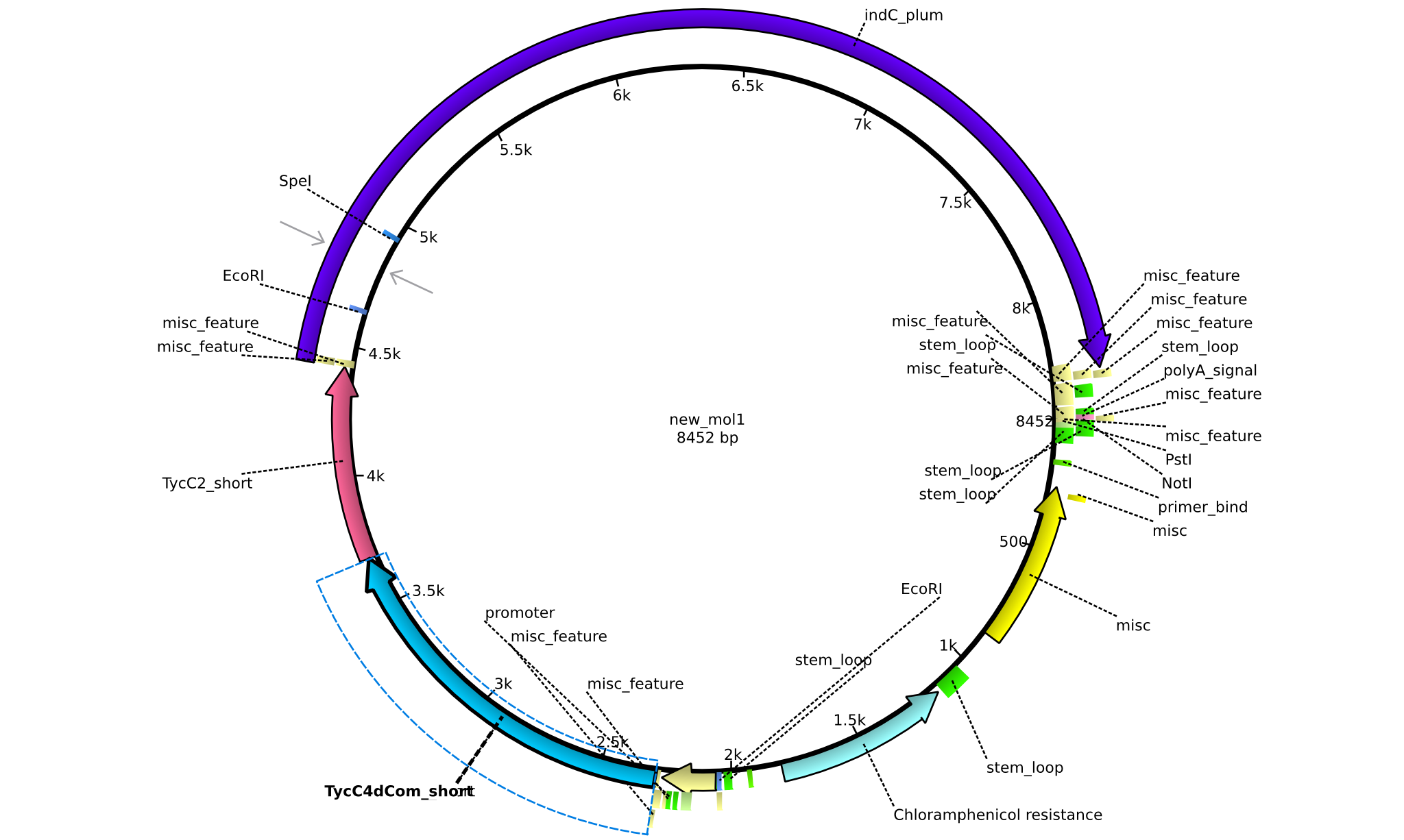 | pAR01 |
| pAR02 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C4 short linker | pSB1C3-TycC4dCom_s+TycC2_m+indC | 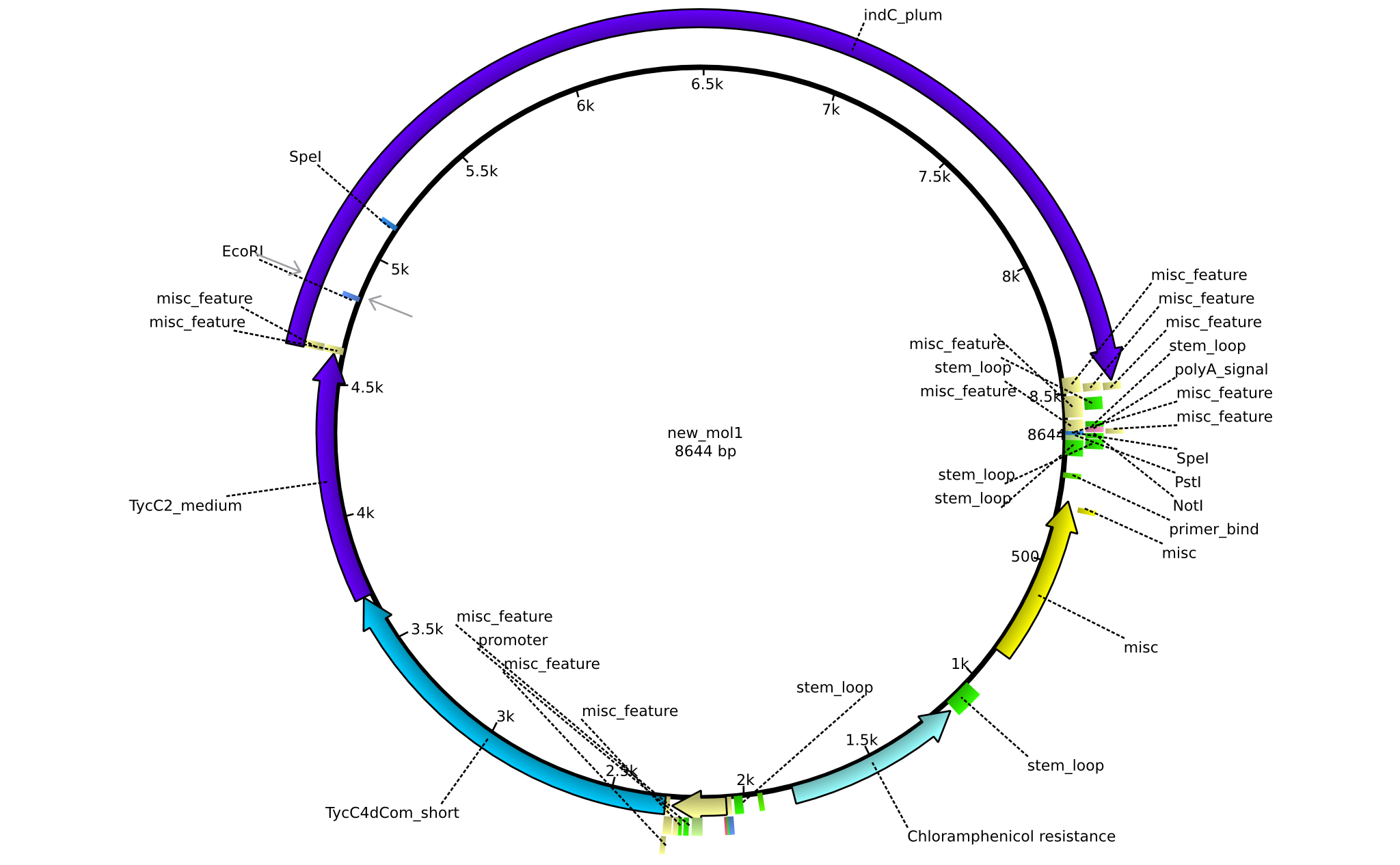 | pAR02 |
| pAR03 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C2 short linker | pSB1C3-TycC4dCom_m+TycC2_s+indC | 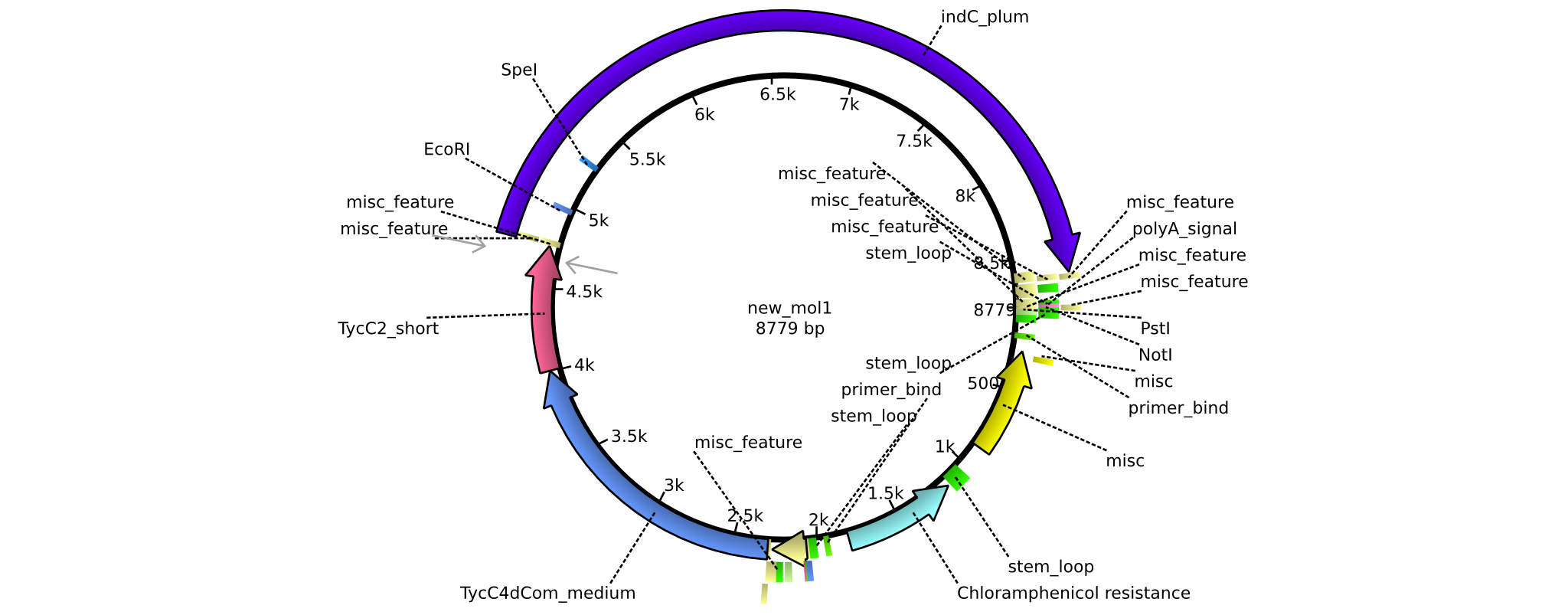 | pAR03 |
| pAR04 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with medium linkers | pSB1C3-TycC4dCom+TycC2+indC | 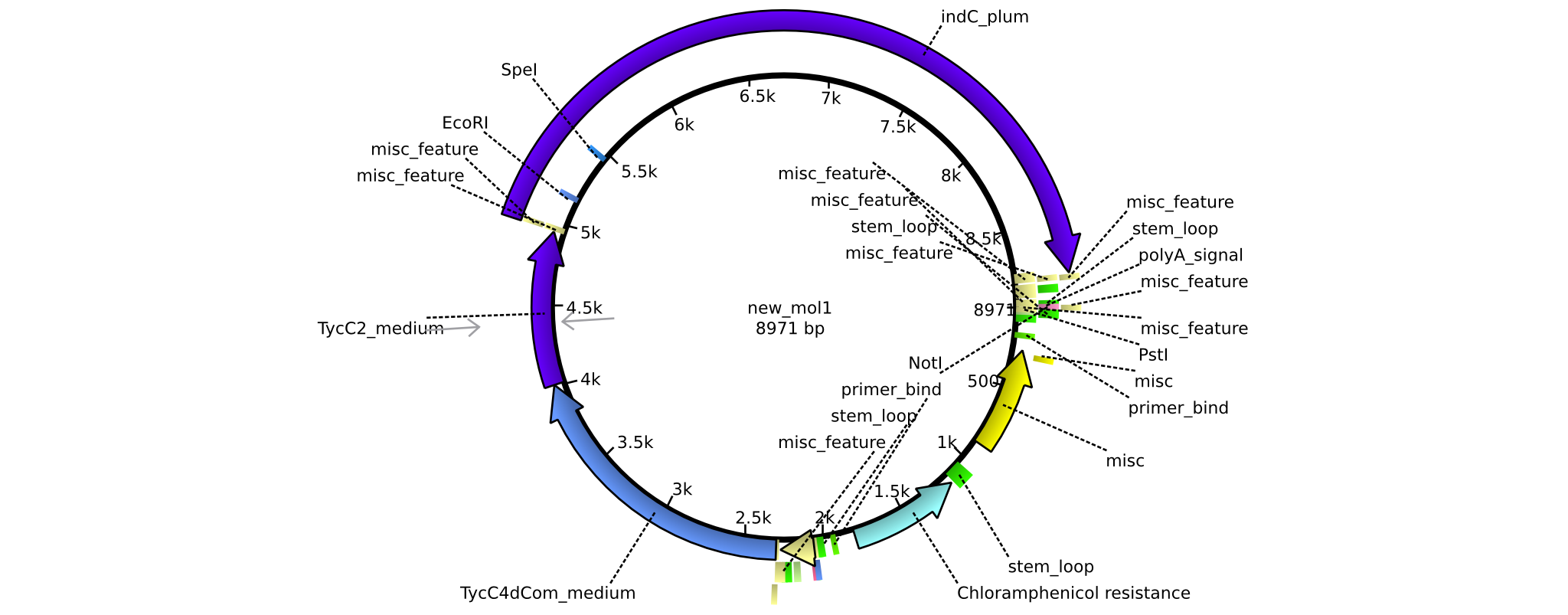 | pAR04 |
| pAR05 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C2 long linker | pSB1C3-TycC4dCom_m+TycC2_l+indC | 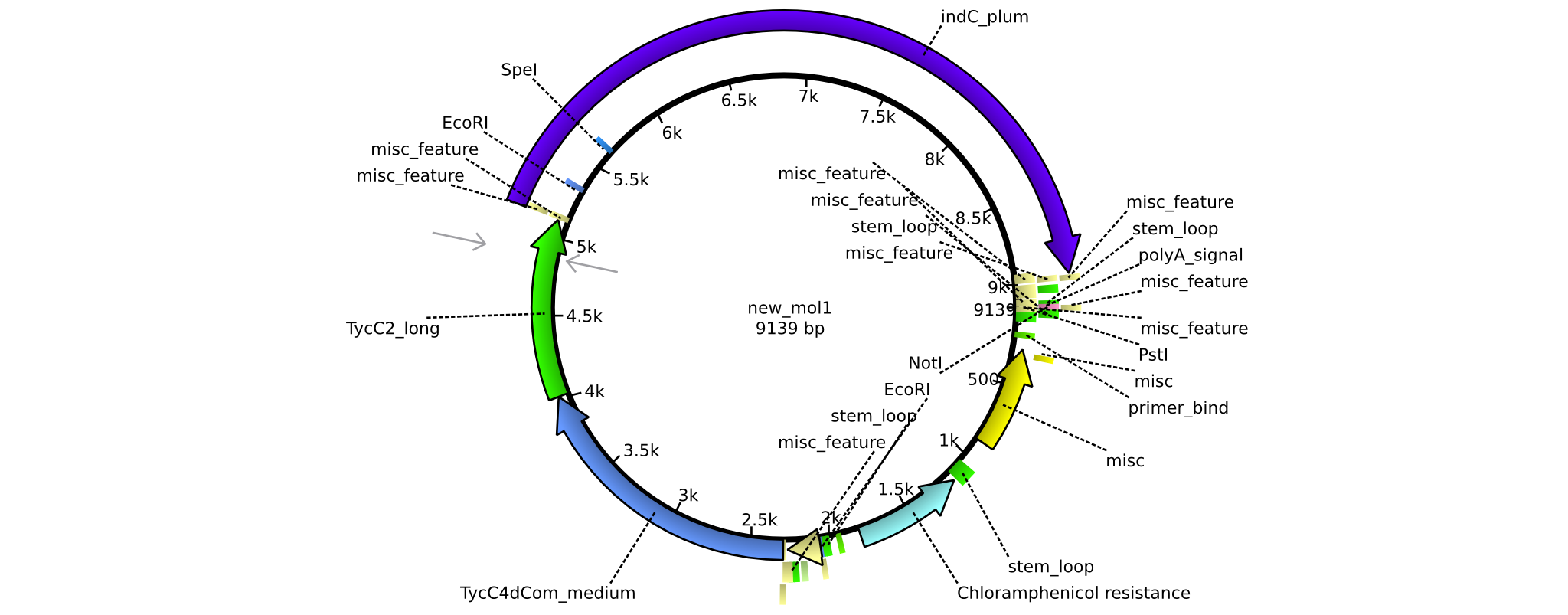 | pAR05 |
| pAR06 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C4 long linker | pSB1C3-TycC4dCom_l+TycC2_m+indC | 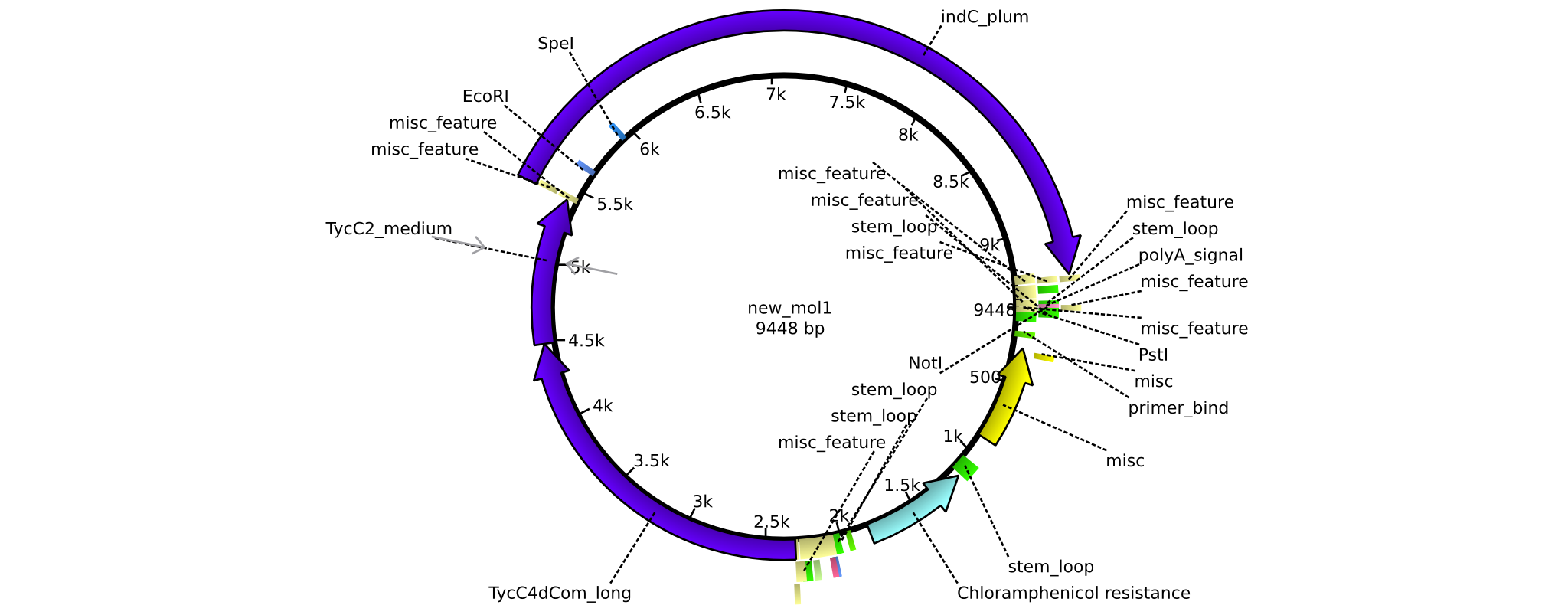 | pAR06 |
| pAR07 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with long linkers | pSB1C3-TycC4dCom_l+TycC2_l+indC | 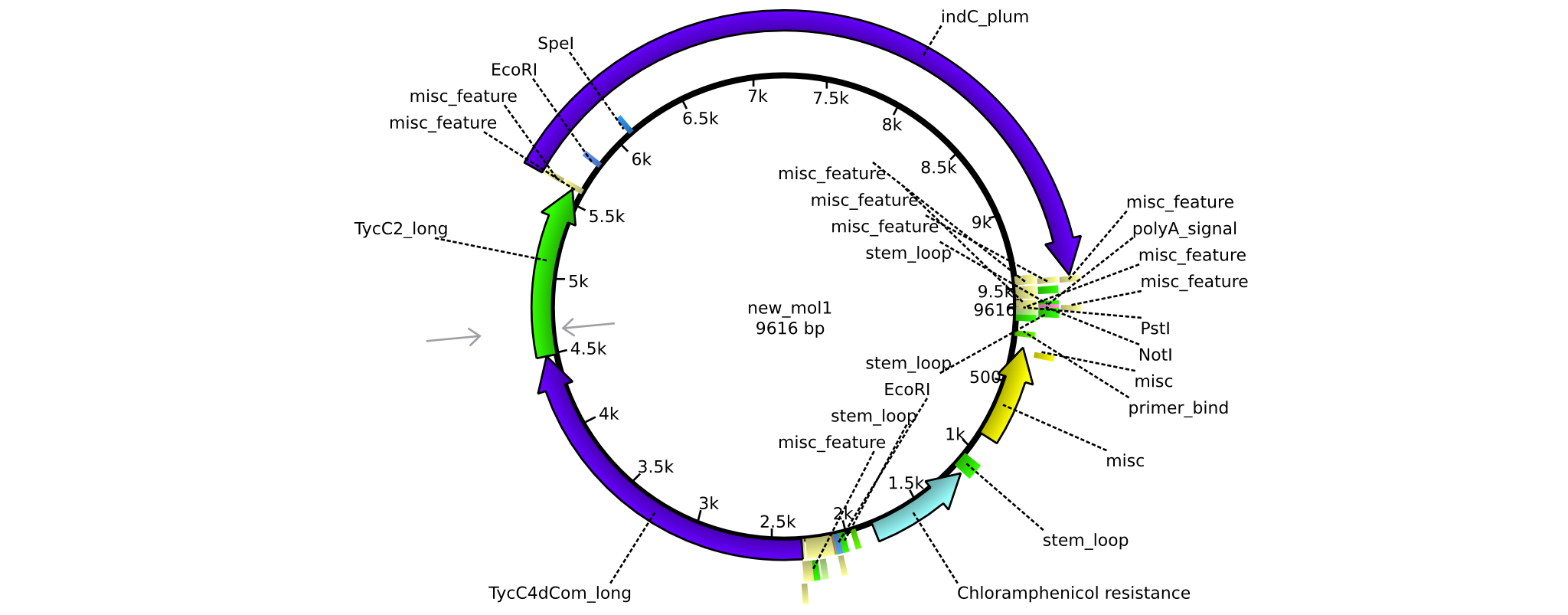 | pAR07 |
| pAR08 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C4 short and C2 long linker | pSB1C3-TycC4dCom_s+TycC2_l+indC | 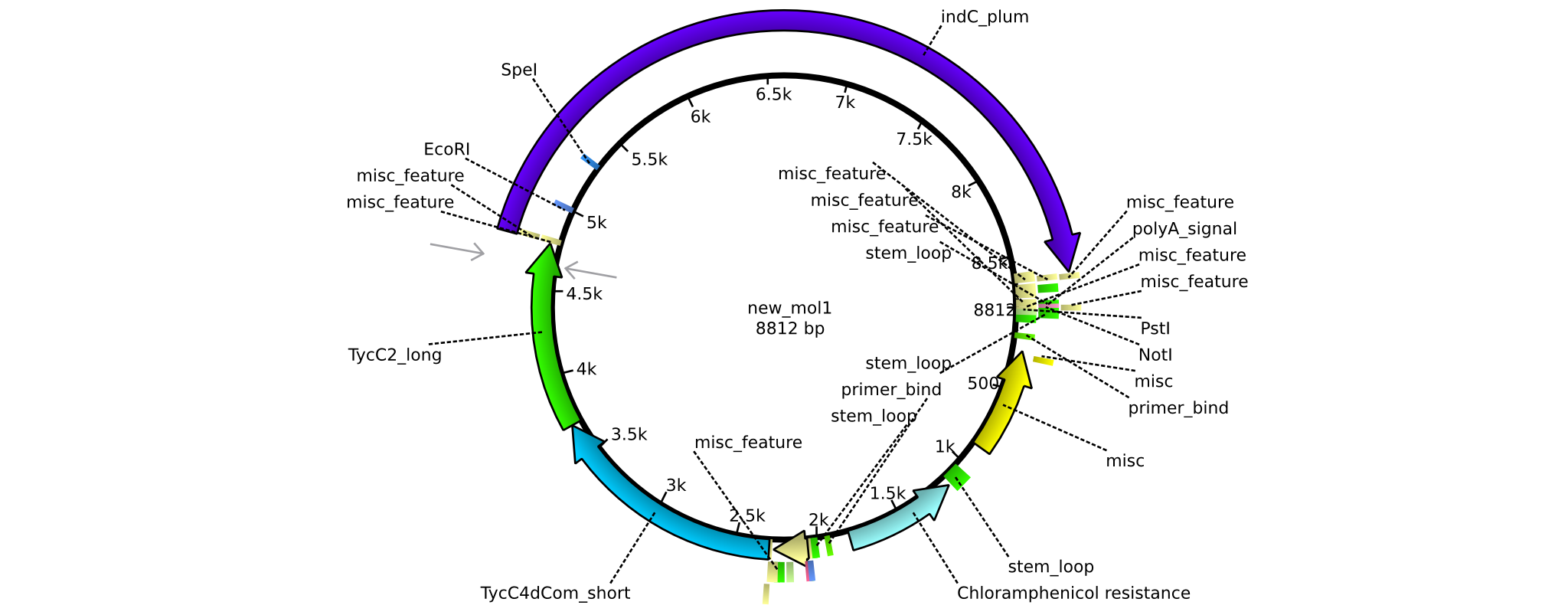 | pAR08 |
| pAR09 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C4 long and C2 short linker | pSB1C3-TycC4dCom_l+TycC2_s+indC | 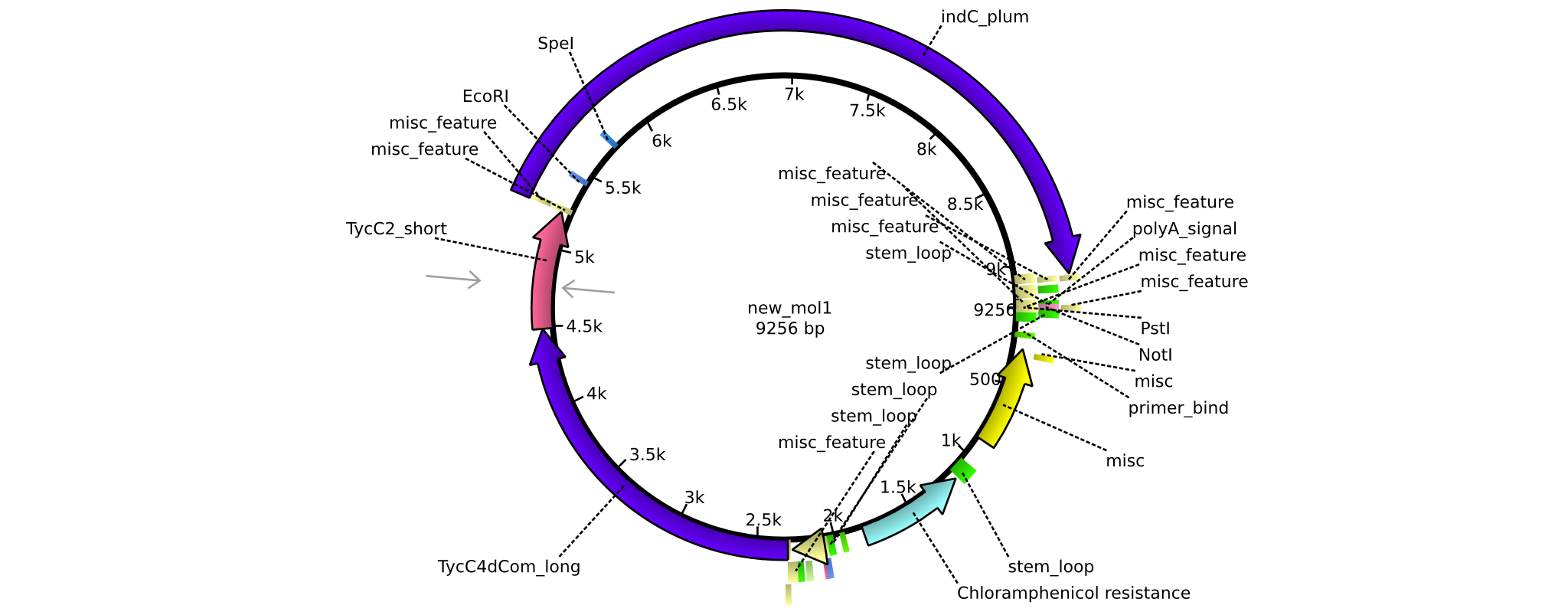 | pAR09 |
Dinner plasmids
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
pIK6 | 2013-08-27 | pIK6: special Ilia plasmid. This plasmid does not fullfill iGEM's RFC10 but is nevertheless special. It is for expression of tasty Ham driven by a strong lacCheese inducible promoter, a RBSalami for increased fleshiness, which can only terminated by a Champignon stop codon. The pIZa backbone with Pepper resistance and high copy Onion ori will improve the users taste stimulation. Bon Appétit! | pIZa-lacCheese P-RBSalami-ham CDS-Champ Term-Pepper res-Onion Ori | 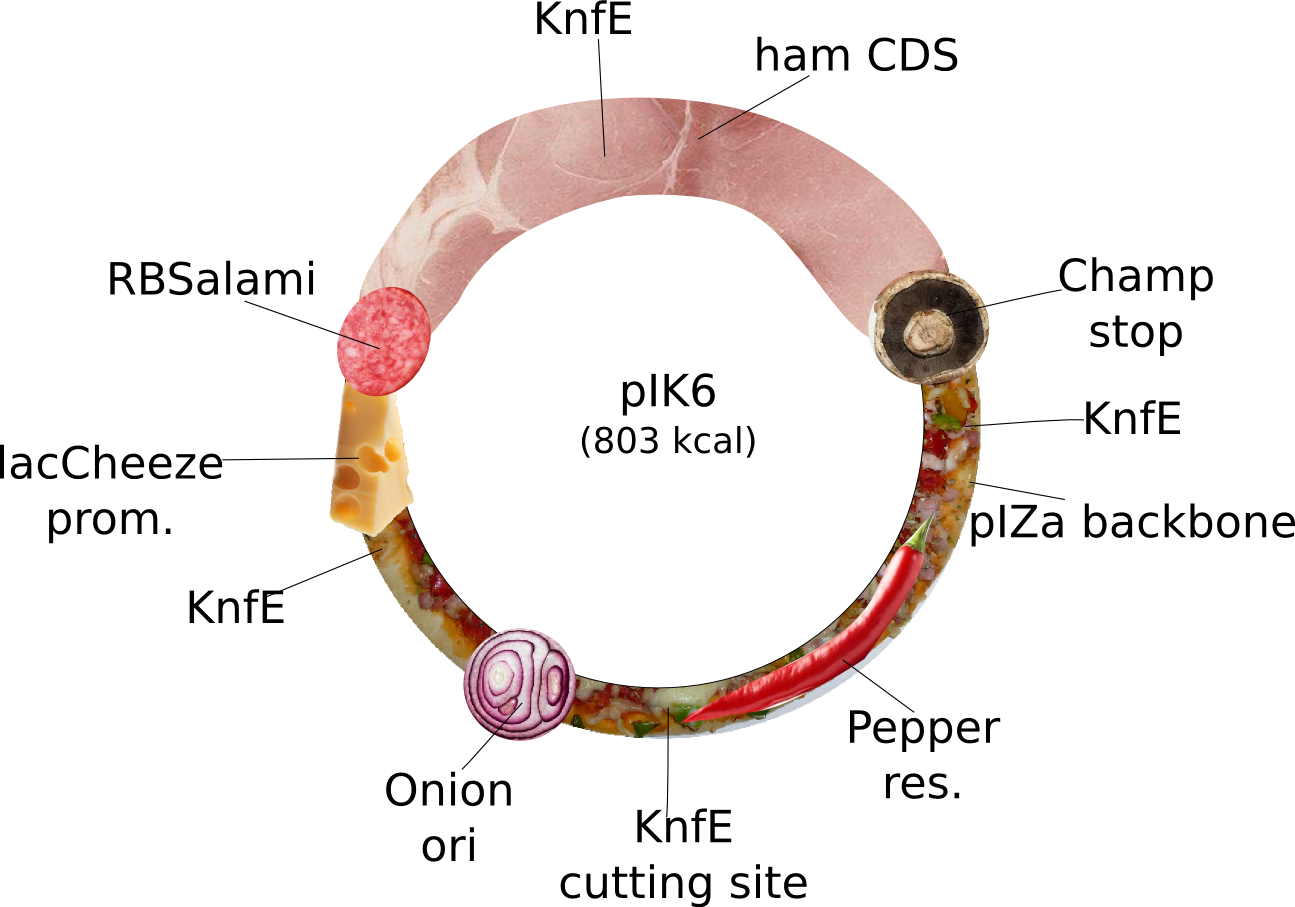 | pIK6 |
Domain Shuffling and PPTases
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
pMM64 | 2013-06-01 | bpsA(fus) | pETDuet-1-T7lac-bpsA(fus) | 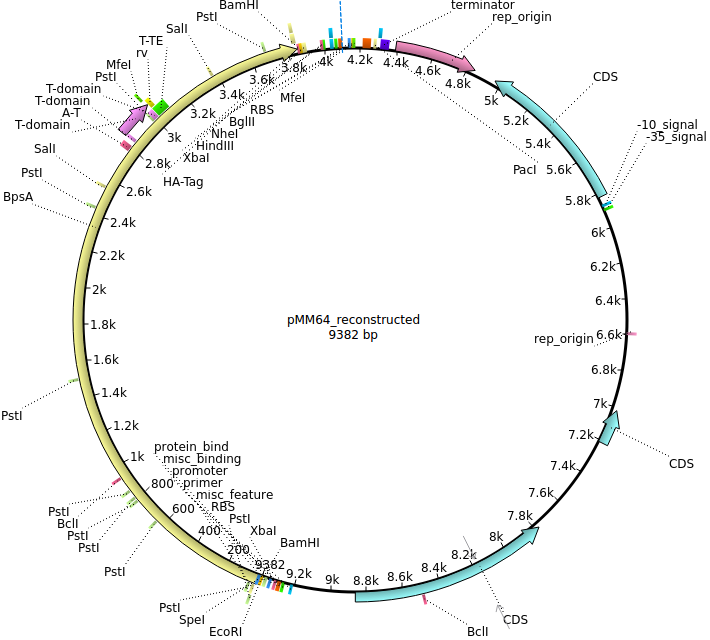 | pMM64 |
pMM65 | 2013-06-01 | svp(fus) | pETDuet-1-T7lac-svp(fus) | 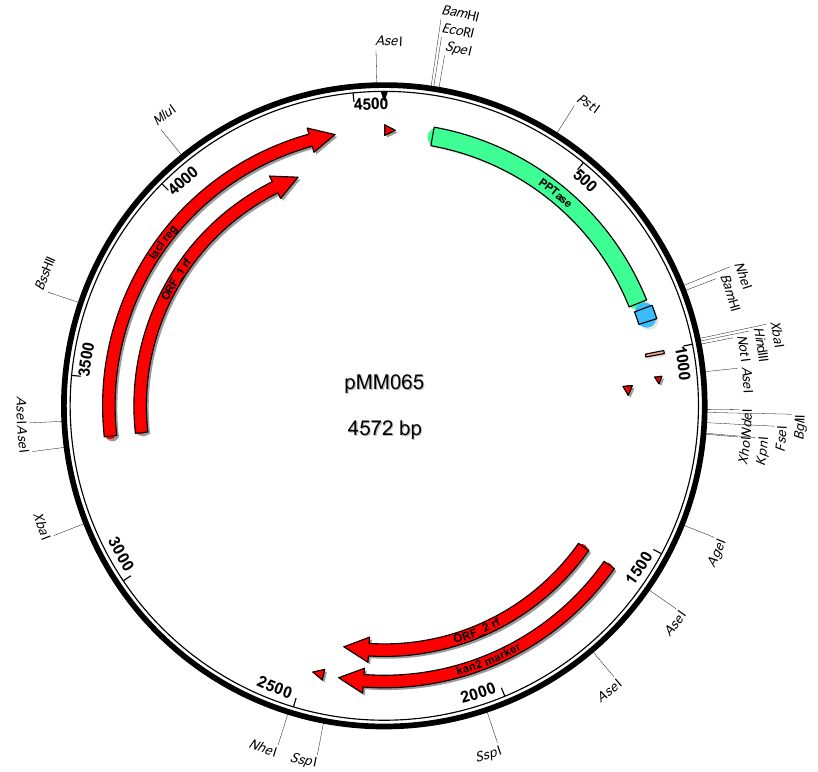 | pMM65 |
pKH4 | 2013-09-01 | pRB3/pMM64 derived plasmid without sfp | pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-nonsense | 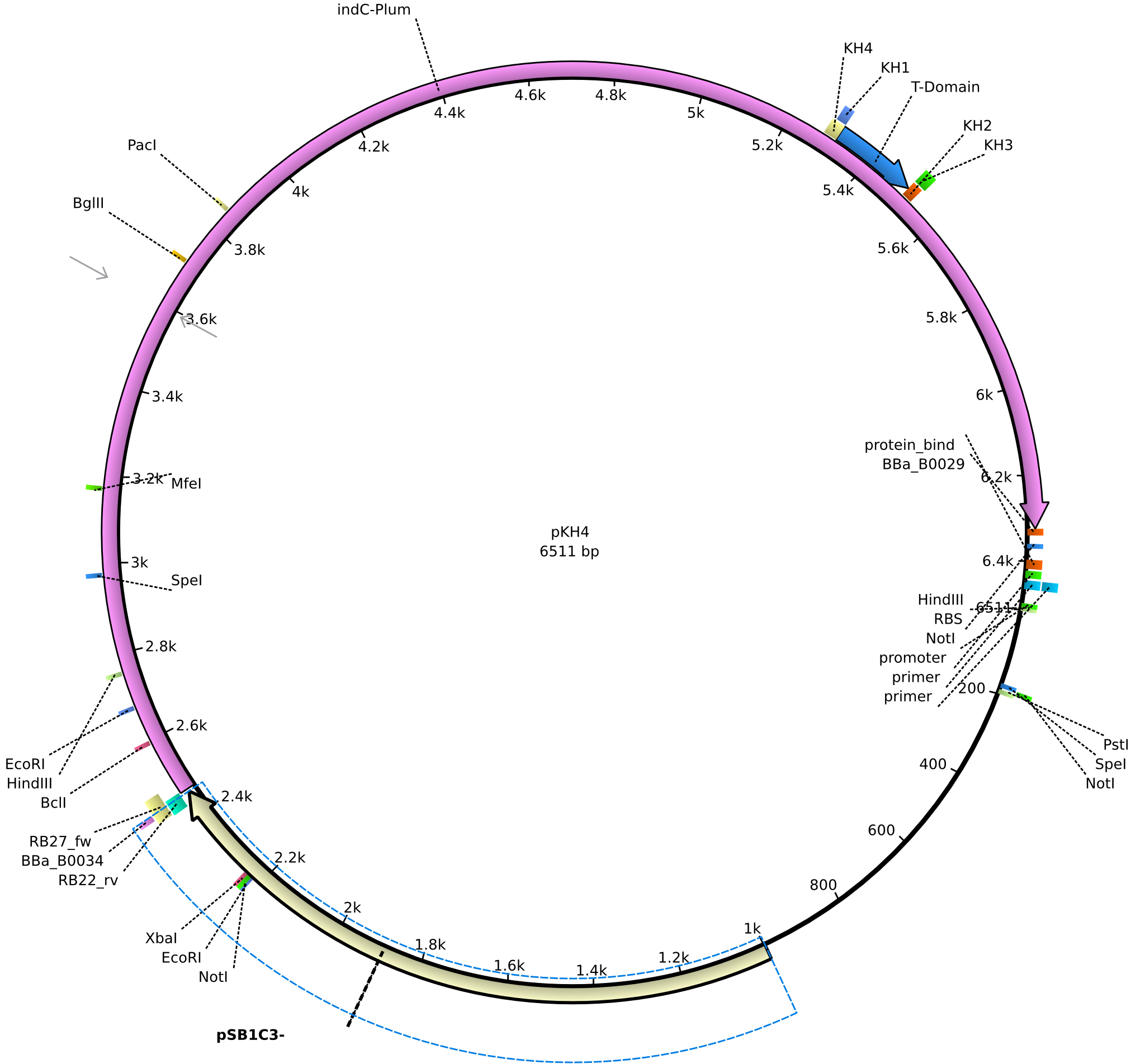 | pKH4 |
pKH6 | 2013-09-05 | BBa CDS sfp only for part submission | pSB1C3-sfp | 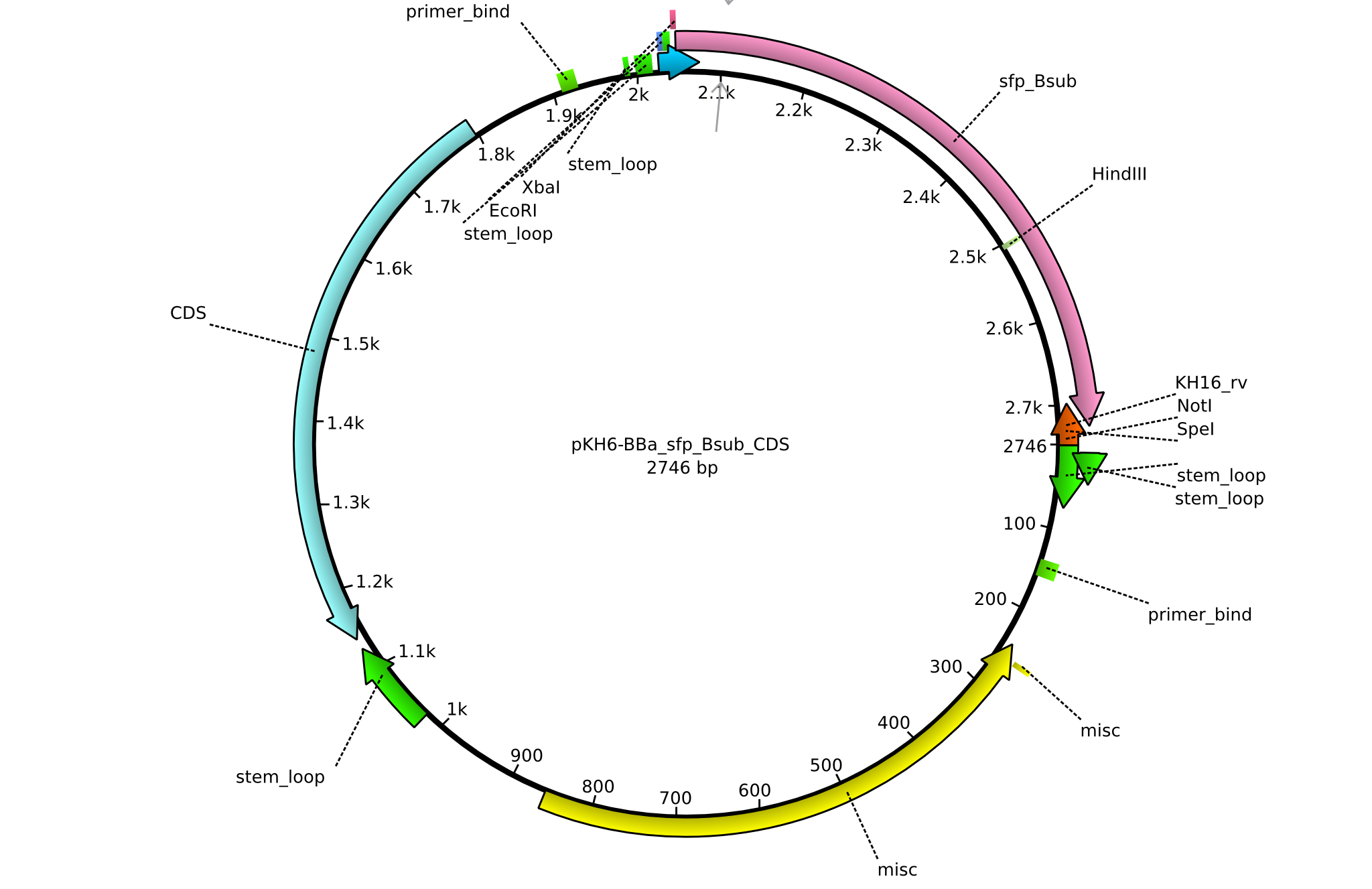 | pKH6 |
pKH7 | 2013-09-05 | BBa CDS svp only for part submission | pSB1C3-svp | 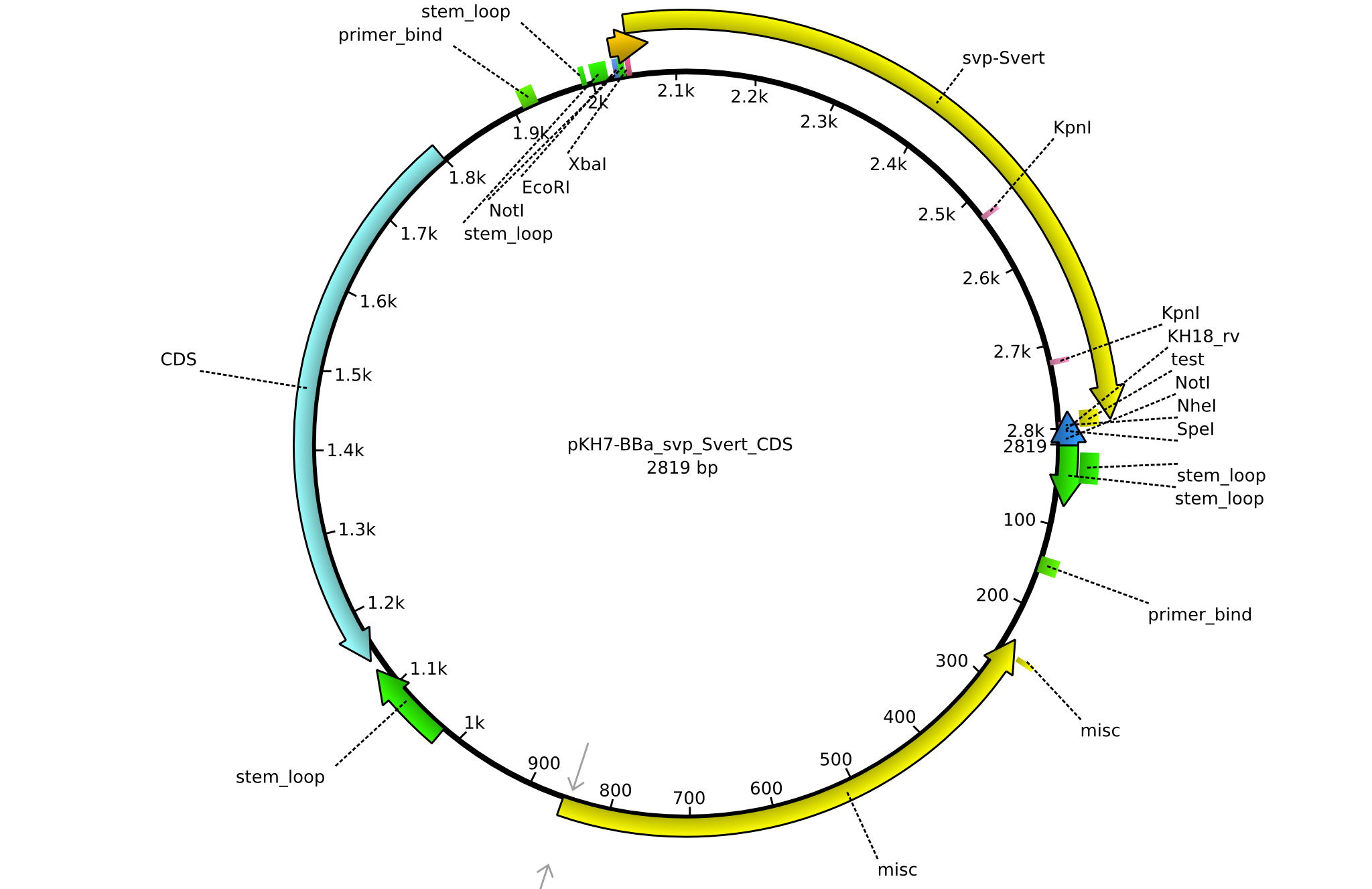 | pKH7 |
pKH8 | 2013-09-05 | BBa CDS entD only for part submission | pSB1C3-entD | 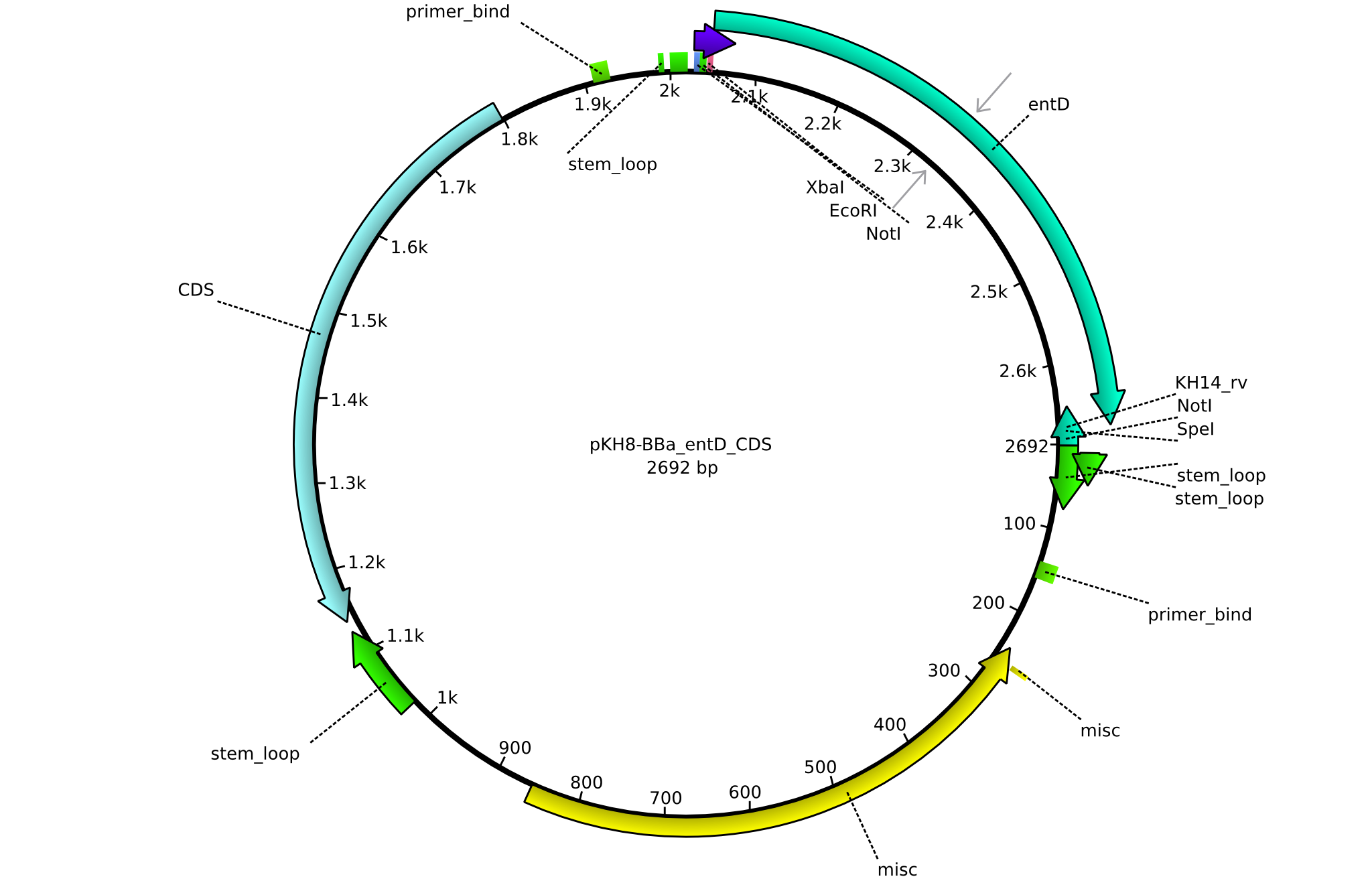 | pKH8 |
pKH9 | 2013-09-05 | BBa CDS delC only for part submission | pSB1C3-delC | 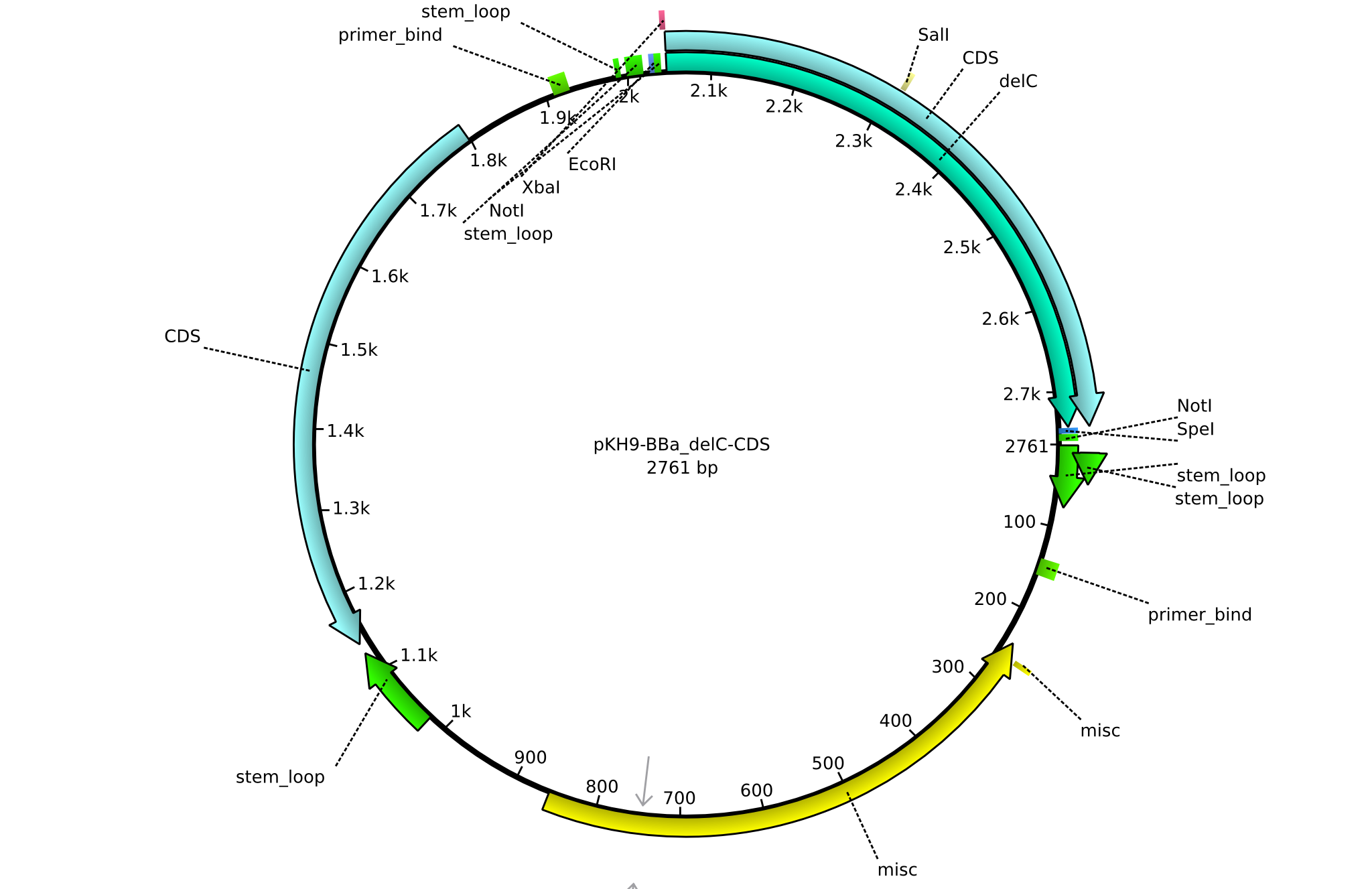 | pKH9 |
pRB1 | 2013-07-01 | bpsA svp(pMM65) | pSB1C3-lacP-BBa_B0034-bpsA-BBa_B0029-svp(pMM65) | pXXYY | pXXYY |
pRB2 | 2013-07-08 | bpsA svp | HindIII-pSB1C3-lacP-BBa_B0034-bpsA-BBa_B0029-HindIII-svp | pXXYY | pXXYY |
pRB3 | 2013-07-15 | indC sfp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-BamHI-sfp | 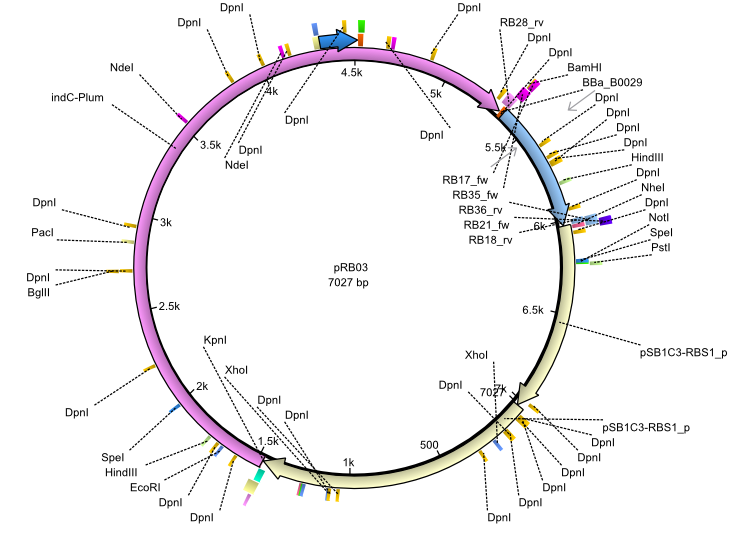 | pRB03 |
pRB4 | 2013-07-15 | indC svp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-BamHI-svp | 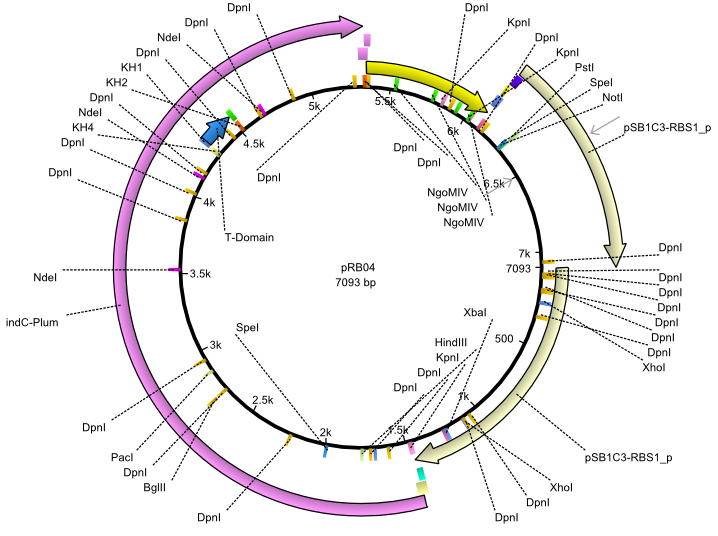 | pRB04 |
pRB5 | 2013-07-15 | indC svpF | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-BamHI-svp(pMM65) | 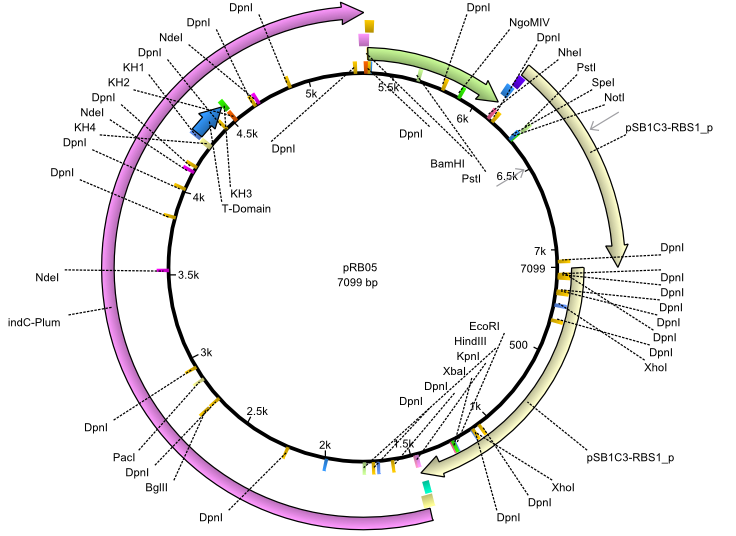 | pRB05 |
pRB6 | 2013-07-15 | indC entD | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-BamHI-entD | 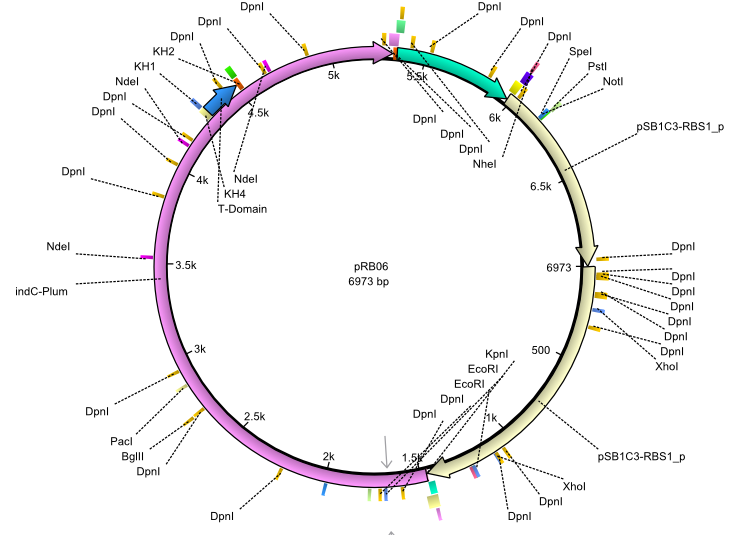 | pRB06 |
pRB7 | 2013-07-15 | bpsA(pMM64) sfp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-bpsA(pMM64)-BBa_B0029-BamHI-sfp | 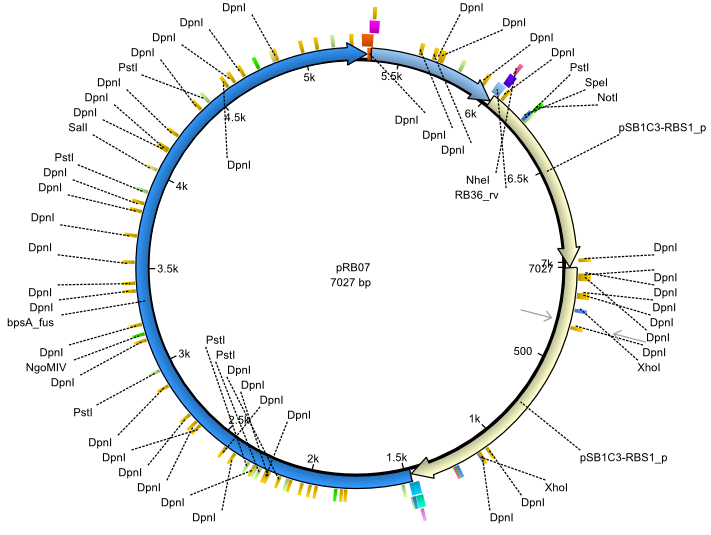 | pRB07 |
pRB8 | 2013-07-15 | bpsA(pMM64) svpF | NheI-pSB1C3-lacP-BBa_B0034-KpnI-bpsA(pMM64)-BBa_B0029-BamHI-svp(pMM65) | 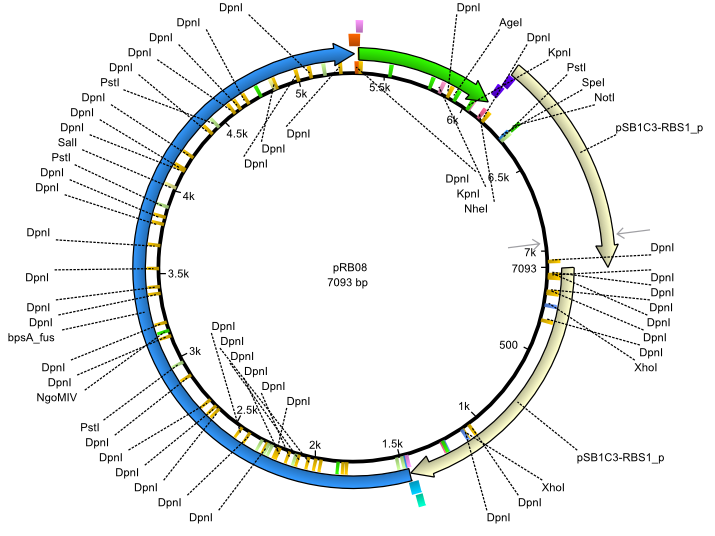 | pRB08 |
pRB9 | 2013-07-15 | bpsA(pMM64) svp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-bpsA(pMM64)-BBa_B0029-BamHI-svp | 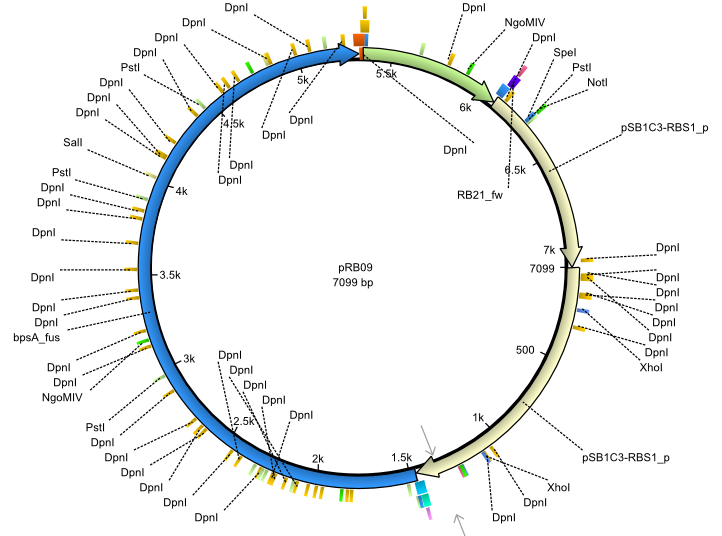 | pRB09 |
pRB10 | 2013-07-15 | bpsA(pMM64) entD | NheI-pSB1C3-lacP-BBa_B0034-KpnI-bpsA(pMM64)-BBa_B0029-BamHI-entD | 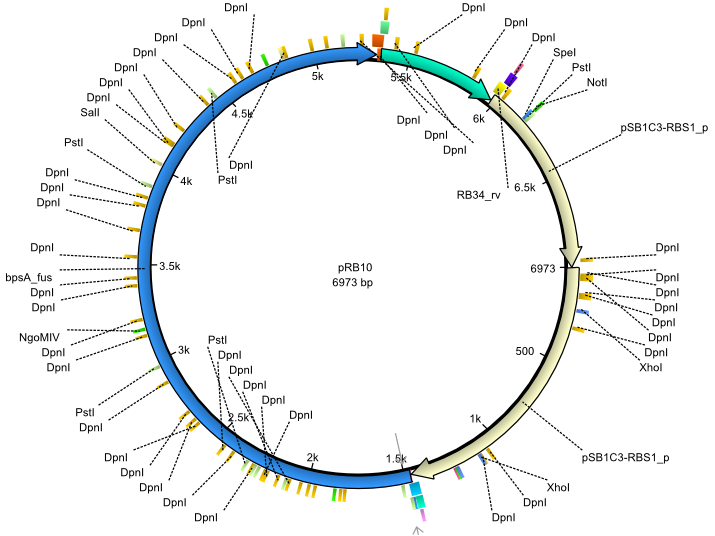 | pRB10 |
pRB11 | 2013-07-29 | pKH1-der bpsA(pMM64)-ccdb svpF | pSB1C3-lacP-BBa_B0034-bpsA(pMM64)(ccdb)-BBa_B0029-svp(pMM65) | pXXYY | pXXYY |
pRB12 | 2013-07-29 | pKH2-der bpsA(pMM64)-ccdb svpF | pSB1C3-lacP-BBa_B0034-bpsA(pMM64)(ccdb)-BBa_B0029-svp(pMM65) | pXXYY | pXXYY |
pRB13 | 2013-07-29 | pRB3-der indC-ccdb sfp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC(ccdb)-BBa_B0029-BamHI-sfp | pXXYY | pXXYY |
pRB14 | 2013-08-12 | pRB3-der indC-ccdB sfp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC(ccdB)-BBa_B0029-BamHI-sfp | pXXYY | pXXYY |
pRB15 | 2013-08-12 | pSB3K3 sfp | pSB3K3-lacP-BBa_B0029-BamHI-sfp-NheI | 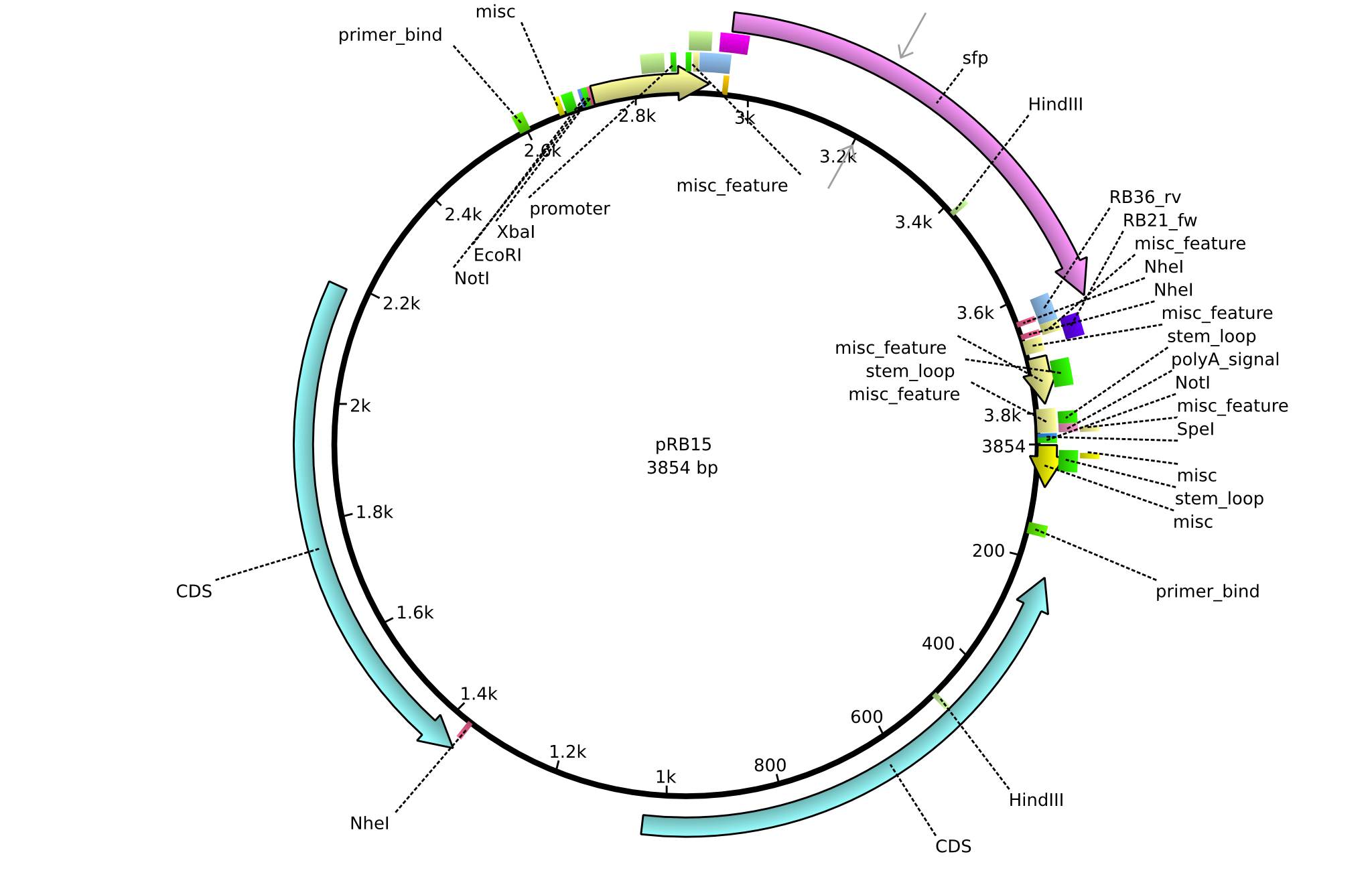 | pRB15 |
pRB16 | 2013-08-12 | pSB3K3 svp | pSB3K3-lacP-BBa_B0029-BamHI-svp-NheI | 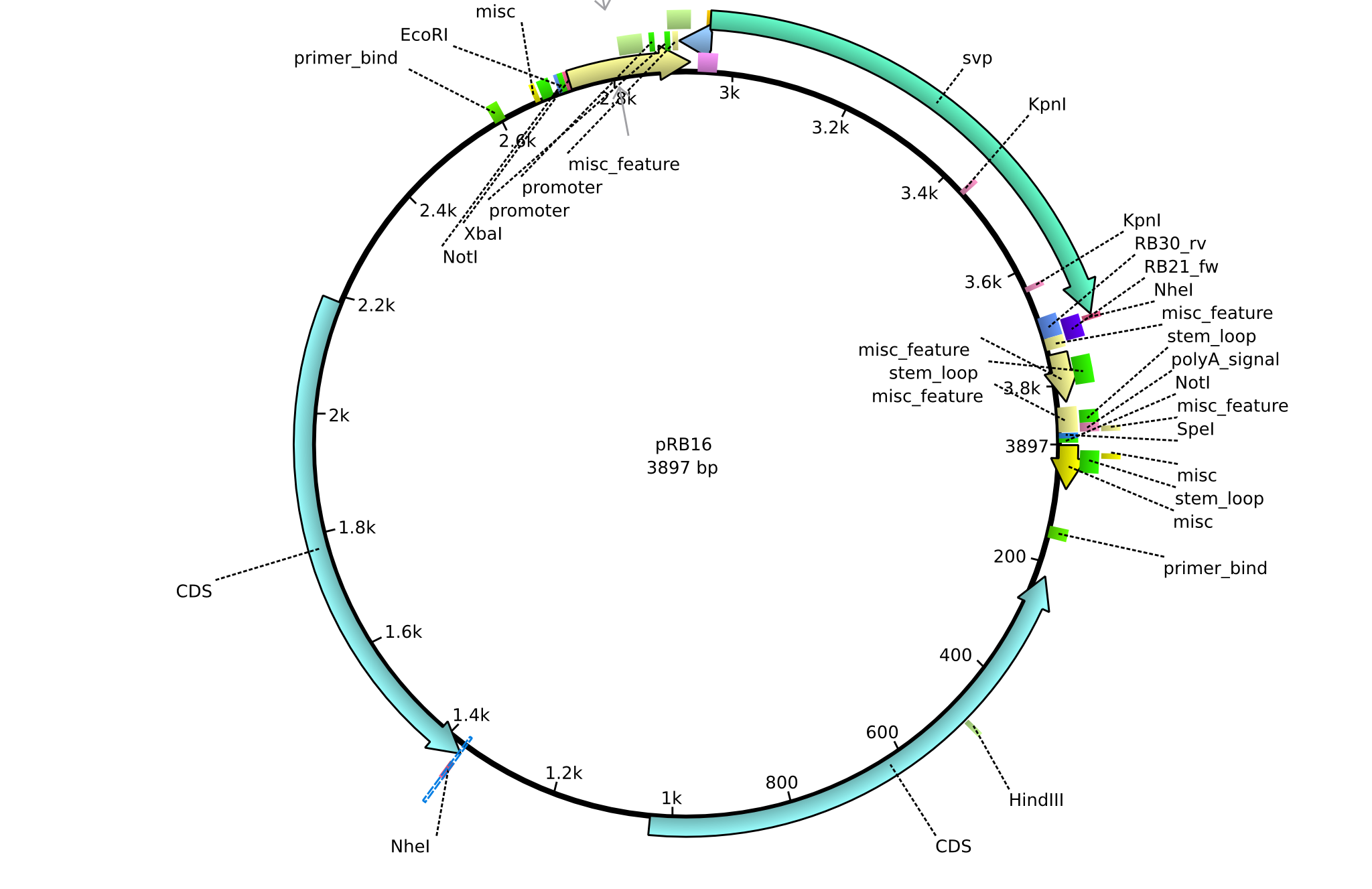 | pRB16 |
pRB17 | 2013-08-12 | pSB3K3 entD | pSB3K3-lacP-BBa_B0029-BamHI-entD-NheI | 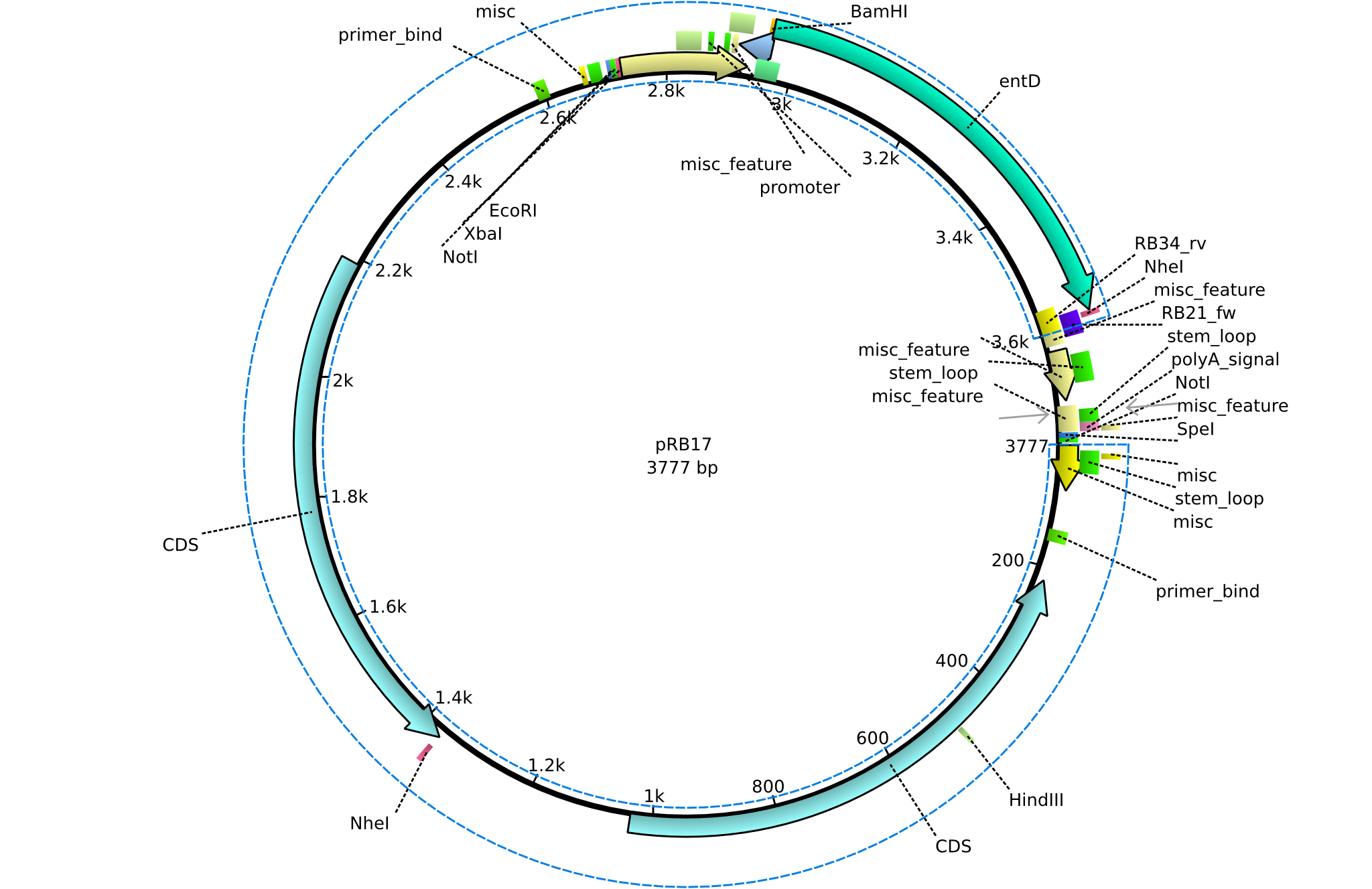 | pRB17 |
pRB18 | 2013-08-12 | pSB3K3 delC | pSB3K3-lacP-BBa_B0029-BamHI-delC-NheI | 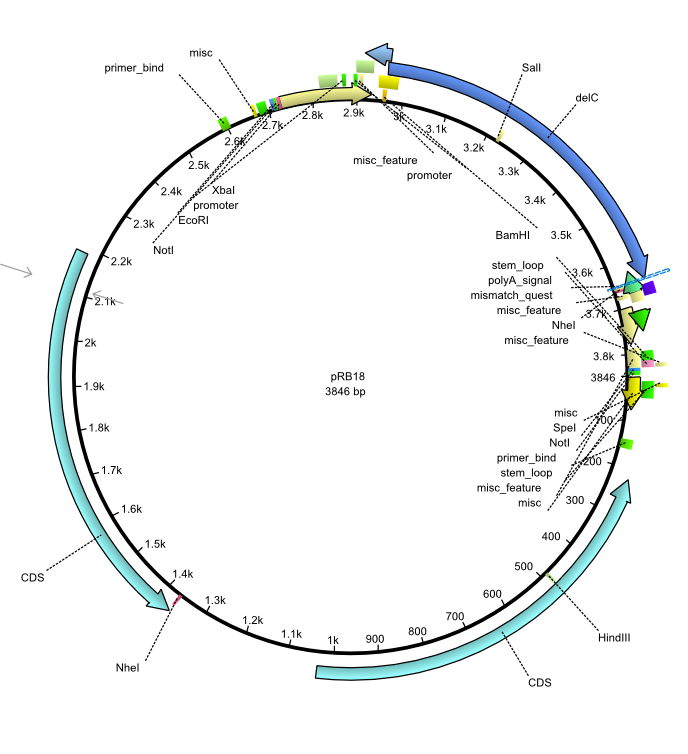 | pRB18 |
pRB19 | 2013-08-19 | pRB14-der indC-ccdB | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC(ccdB) | pXXYY | pXXYY |
pRB20 | 2013-08-19 | pRB19-der indC-HD-ccdB | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-HD(ccdB) | pXXYY | pXXYY |
pRB21 | 2013-08-19 | pSB1C3 indC | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC | 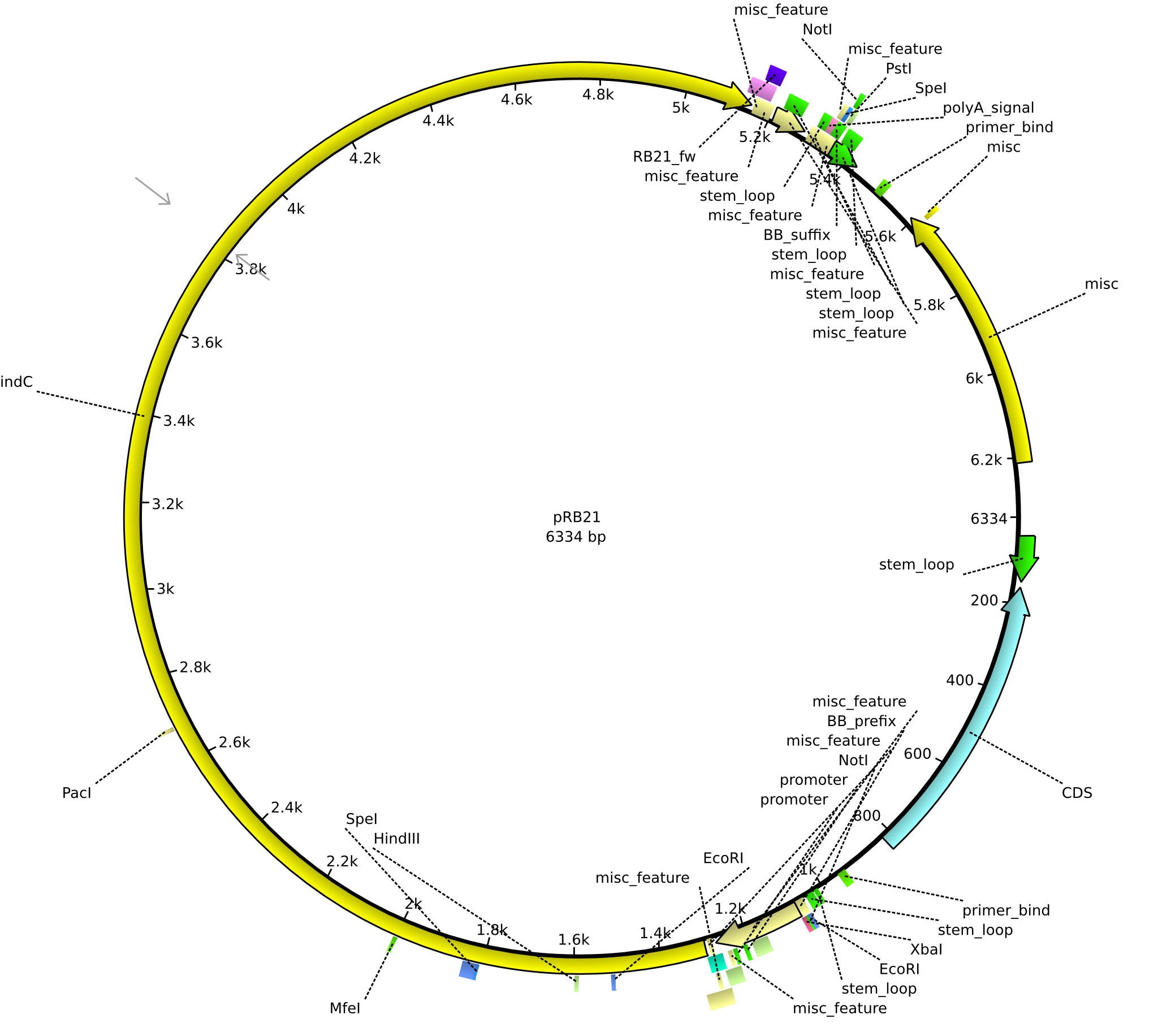 | pRB21 |
pRB22 | 2013-08-19 | pRB21-der indC-HD | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-HD | 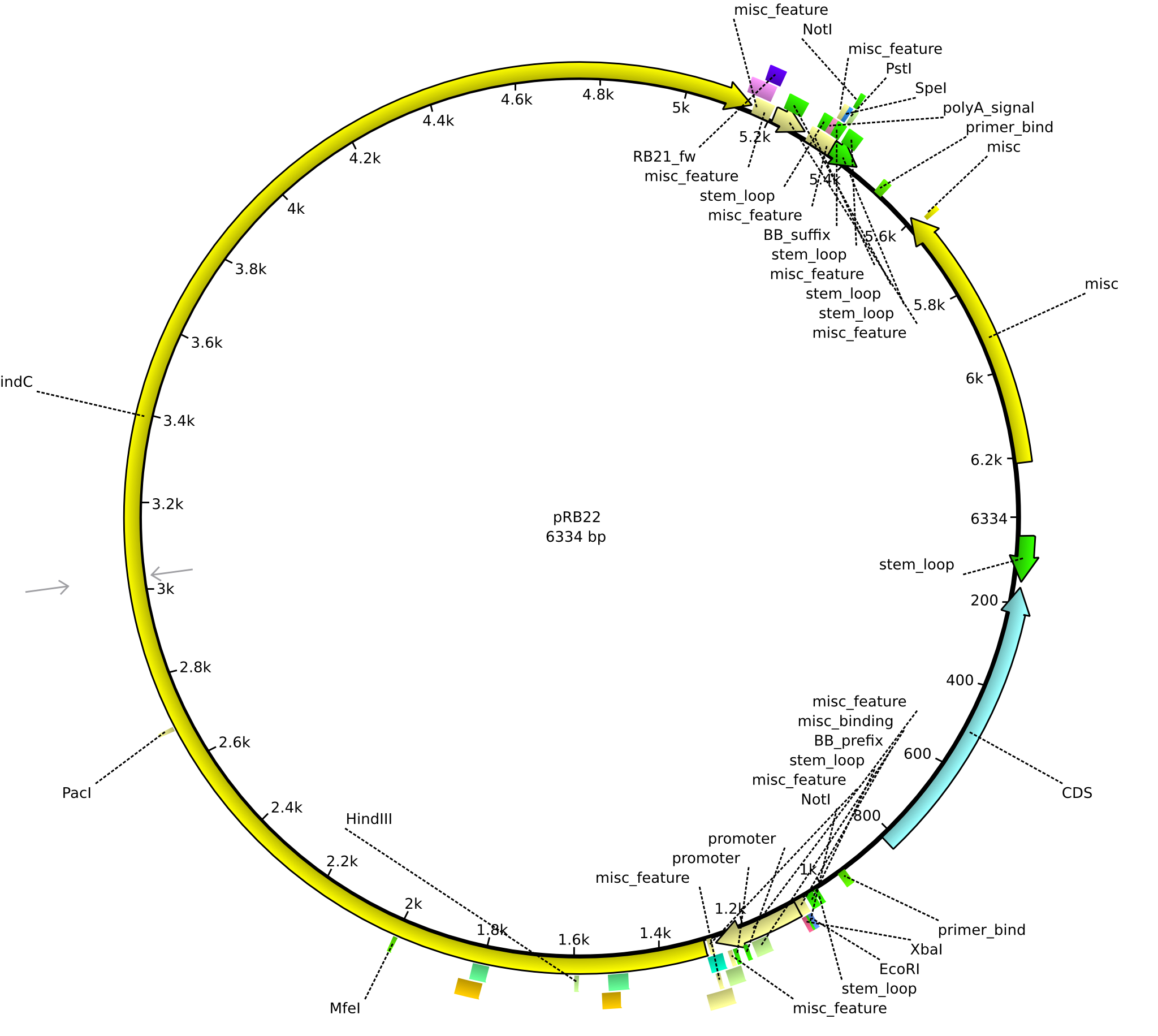 | pRB22 |
pRB23 | 2013-08-19 | pRB22-der indC-HD | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC*dT(ccdB) | pRB23 | pRB23 |
| ID | Name (derived from) | Description |
|---|---|---|
-T1 | indC | native T-Domain of indC from P. luminescens |
-T2 | bpsA | native T-Domain of bpsA from S. lavendulae |
-T3 | entF | native T-Domain of entF from E. coli |
-T4 | tycA1 | native T-domain of tycA1 from Brevibacillus parabrevis |
-T5 | tycC6 | native T-domain of tycC6 from B. parabrevis |
-T6 | delH4 | native T-domain of delH4 from Delftia acidovorans |
-T7 | delH5 | native T-domain of delH5 from D. acidovorans |
-T8 | plu2642 | native T-domain of plu2642 from Photorhabdus luminescens |
-T9 | plu2670 | native T-domain of plu2670 from P. luminescens |
-T10 | synT1 | synthetic T-Domain from indC-BLAST |
-T11 | synT2 | synthetic T-Domain from indC_T-Plum-BLAST |
-T12 | synT3 | synthetic T-Domain from indC_T-BLAST |
-T13 | synT4 | synthetic T-Domain from project_5 BLAST |
-T14 | synT5 | synthetic T-Domain random 1 (indigoidine synthetases) |
-T15 | synT6 | synthetic T-Domain random 2 (project_5) |
-TTE1 | bpsA | native TTE-Domain of bpsA from S. lavendulae |
-TTE2 | entF | native TTE-Domain of entF from E. coli |
-TTE3 | tycC6 | native TTE-domain of tycC6 from B. parabrevis |
-TTE4 | delH5 | native TTE-domain of delH5 from D. acidovorans |
Acidovorans complex medium
for 1 L
- 0.5 g yeast extract (Difco-BectonDickinson)
- 1.0 g Cas amino acids (Difco)
- 2.0 g pyruvic acid
- 2.0 g L-glutamine
- 0.3 g KH2PO4
- 0.3 g MgSO4
- 2.0 g MOPS
- 4.0 g Chelex 100-resin (Sigma)
- pH adjusted to 7.2–7.3 with 5 M KOH
- fill up to 900mL before adding pyruvic acid and L-glutamine
- adjust pH
- fill up to 1L
- treat with Chelex for 1h
- remove Chelex by filtration
Ampicillin Stock Solution
| What | Ampicillin 100 mg / ml |
| Amount | 50 ml |
| Storage | -24°C freezer |
| Notes | Use in 1:1000 dilution |
Efficiency test
Because we daugthed the effiency of our stock solution, we prepared a little effiency test. TB or LB media were prepared with different ampicillin solutions in order to detect at which concentration bacteria cells carrying a mRFP expressing plasmid loose it.


We can conclude that our ampicilin should still be usable, since it is at least as efficient as the ampicilin stock solution optained from a different group.
Bacitracin stock solution
| What | Bacitracin 5000 U/ml |
| Amount | 5 ml |
| Storage | -24°C freezer |
Brevibacillus parabrevis glycerol stock
| What | Brevibacillus parabrevis ATCC8185 glycerol stock |
| Amount | 16 x 1.3 ml |
| Storage | -80°C freezer |
Chloramphenicol stock solution
| What | Chloramphenicol 30 mg / ml |
| Amount | 10 ml |
| Storage | -24°C freezer |
| Notes | Solved in 100% ethanol. Use in 1:3000 dilution |
D. acidovorans glycerol stock
| What | D. acidovorans glycerol stock |
| Amount | 10 x 1 ml |
| Storage | -80°C freezern |
BAP 1 glycerol stock
| What | E. coli BAP1 glycerol stock |
| Amount | 2 x 1.3 ml |
| Storage | -80°C freezer |
E.coli BAP1, competent
| What | E. coli BAP1 |
| Amount | 18 x 100 µl |
| Storage | -80°C freezer |
| Notes | Grows extremely fast. Be careful with miniPreps, at least in cultures with ampicillin it tends to degrade all available ampicillin and then lose the respective plasmid. |
E. coli BAP1-pKD46 glycerol stock
| What | E. coli BAP1-pKD46 glycerol stock (Ampr) |
| Amount | 2 x 1.3 ml |
| Storage | -80°C freezer |
| Notes | Grow at 30°C only! Growth at 37°C will lead to loss of pKD46 plasmid. |
BAP1-pLF03 glycerol stock
| What | E. coli BAP1-pLF03 glycerol stock (Ampr) |
| Amount | 1 x 1.3 ml |
| Storage | -80°C freezer |
| Notes | Might have low amount of plasmid-carrying bacteria due to long culturing time (all Amp in medium cleaved) |
E. coli DH5α-pCP20 glycerol stock
| What | E. coli DH5α-pCP20 glycerol stock (Ampr, Cmr) |
| Amount | 2 x 1.3 ml |
| Storage | -80°C freezer |
| Notes | Grow at 30°C only! Growth at 37°C will lead to loss of pCP20 plasmid. |
Heat-shock competent E. coli TOP10
| What | E. coli TOP10, Heat-shock competent |
| Amount | 200 x 100 µl |
| Storage | -80°C freezer |
| Notes | Verified on 2013-06-07. |
E. coli TOP10-BBa I746200/pSB1AK3 glycerol stock
| What | E. coli TOP10-(BBa I746200 in pSB1AK3) glycerol stock (Ampr, Kanr) |
| Amount | 1 x 1.3 ml |
| Storage | -80°C freezer |
 "
"
