Team:Paris Saclay/Project
From 2013.igem.org
Contents |
What are PCBs?
PolyChlorinated Biphenyls (PCBs), are a family of man-made aromatic organic chemicals which consist of a phenyl ring and more or less chlorines. The first PCB was synthesized in 1881 and were developed in the 1940’s. The production of PCBs peaked in 1970’s, but after discovering that PCBs harm human health, their the production of PCBs has been banned in 1979 in U.S. And in France, the total interdict of production and usage of PCBs declared valid in 1987. The Stockholm Convention on Persistent Organic Pollutants (POPs) was signed in 2001, which aims to eliminate or restrict the production and usage of PCBs and other persistent organic pollutants. This world-wide ban of PCBs effective from May 2004.
Detection and degradation of PCB system in E.coli
Since the second half of the XXth century scientists are fully aware of the fact that some species of bacteria living in mediums with high concentrations of PCB are able to degrade the PCB in pyruvate and acetyl CoA easily metabolized by these organisms.
These species structure biofilms were there are regions with variables concentration of
oxygen, decreasing with depth to surface. The bacteria living in this habitat have in most of
case different degradation pathways namely aerobic or anaerobic depending on the spatial
disposition in the biofilm.
Bacteria in aerobic conditions use an aerobic degradation pathway, the PCB oxidative
degradation, and these in anaerobic condition the PCB reductive dechlorination; no one can
use both pathways so as to degrade the PCB.
The reductive dechlorination can reduce the number of chlorines in high chlorinated PCB
making them assimilable by the oxidative degradation, only efficient with low chlorinated
PCB. That’s the reason why these different species coexist in the biofilms.
So our goal in this project is the creation of an organism able to first detect PCB and after
employ a sequential degradation of the PCB using both combined pathways.
For our experiences we used bacteria present in nature that are able to detect and degrade the
'PCB namely Burkholderia xenovorans, Pseudomonas pseudoalcaligenes KF 707,
Rhodococcus jostii RHA1.'
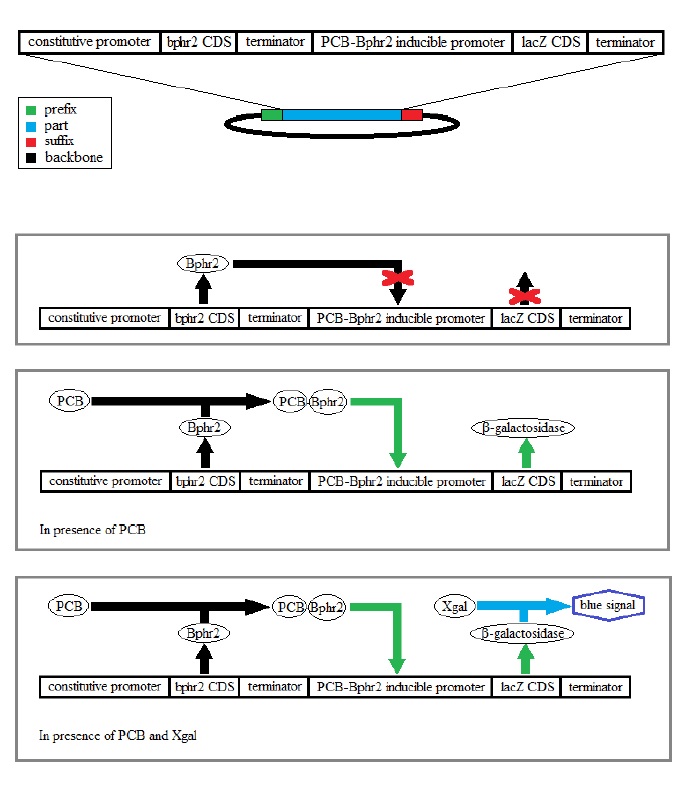
Detection and report of the PCB
In nature these bacteria have a system for the regulation of the oxidative degradation of PCB. This one is based on two regulatory proteins namely Bphr2 and Bphr1 coded respectively by the genes of the same name bphr2 and bphr1.
Bphr2 is able to detect PCB that induces a modification of the protein conformation activating
the beginning of the gene cluster coding for the enzymes doing the oxidative degradation but
also the gene coding for the Bphr1 protein.
The Bphr1 protein can detect the HO-PCB a metabolite derived from PCB, a product of the
beginning of oxidation reactions. In presence of OH-PCB it induces his own transcription and
also the following genes from the cluster that will completely degrade the PCB.
For our construct we will pick out the bphr2 gene and the promoter of the bphr1 gene induced
by PCB-Bphr2 from our species. We will combine the bphr2 coding sequence with a
constitutive promoter that makes up the detection system and finally we will combine the
Bphr1 promoter with the lacZ gene coding for the β-galactosidase enzyme so as to do a
chemical dosing with Xgal and report the signal.
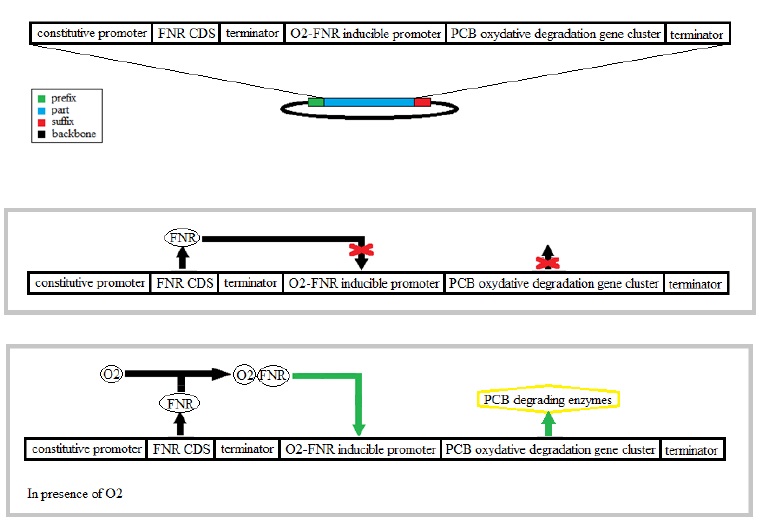
Combination of the aerobic and anaerobic PCB degradation pathways
The bacteria E.coli has an aerobic and an anaerobic metabolism that’s why we used it for the combination of the two degradation pathways. The regulation between pathways in these two conditions is normally made by regulatory proteins like FNR. The FNR protein modifies its conformation in presence of oxygen having an activator or an inhibitor function.
The reductive dechlorination pathway is not well characterized only an enzyme, a
dehalogenase, is mentioned as contributing to this pathway. In these anaerobic conditions the
chlorine takes the place of the oxygen as the electron acceptor.
That’s why we have chosen an activator FNR in presence of oxygen in order to activate the
oxidative degradation.
Team project descriptions are due August 9. The description is only a preliminary description - it will not be used to judge your project. What you write will only serve to provide some background on what your team has been working on so far and what you hope to accomplish.
Description requirements:
- Describe your project on the front page of your team's wiki* or on another page that is easily reached.
- The description only needs to be a couple of paragraphs long.
*Note: The project description does not need to be emailed to iGEM HQ.
 "
"