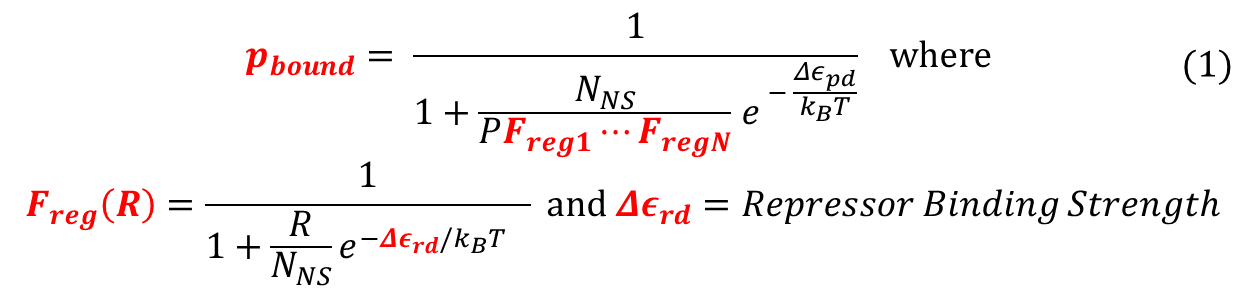Team:Duke/Modeling/2 1
From 2013.igem.org
Contents |
Mathematical Modeling of Bistable Toggle Switch
Application of the Thermodynamic Model of Cooperative Repression
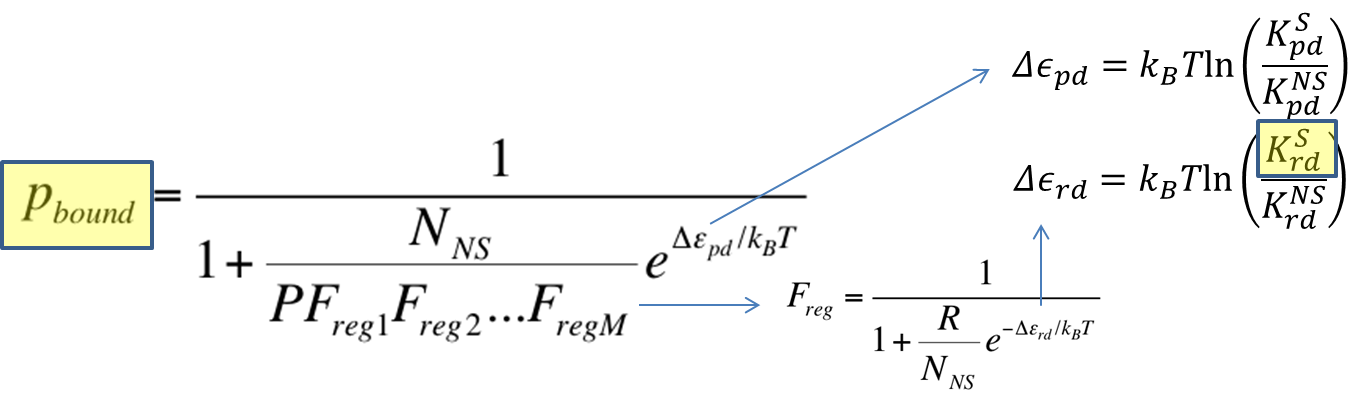
To recap the derivation from the previous section, the key equation for thermodynamic model of cooperative repression is:
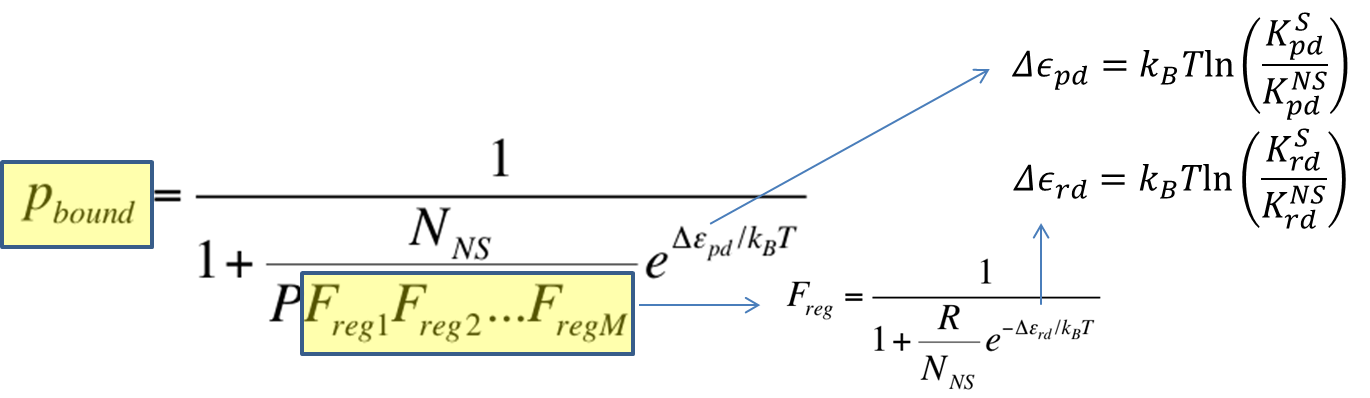
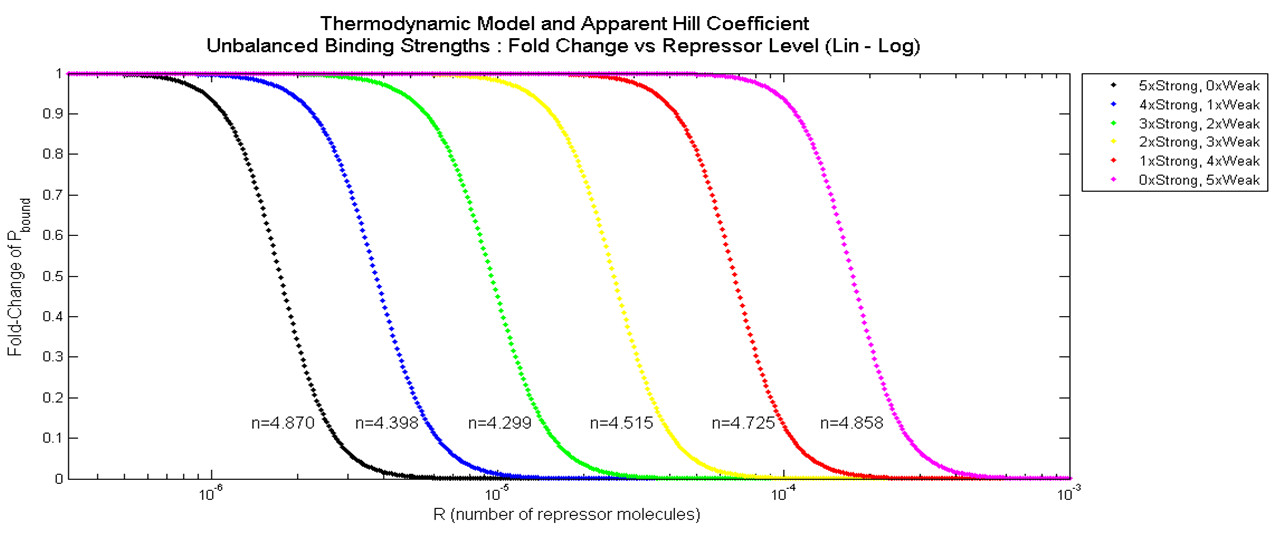
In this section, the above model will be explored; specifically, the relationship between the red-colored terms will be studied. First, the relationship between P_bound and the number of F_reg's will show the effect of changing number of binding sites on level of gene expression. Then, the relationship between P_bound and Ks_rd (Dissociation constant of repressor bound to DNA) will show the effect of changing binding strengths of repressors on level of gene expression.
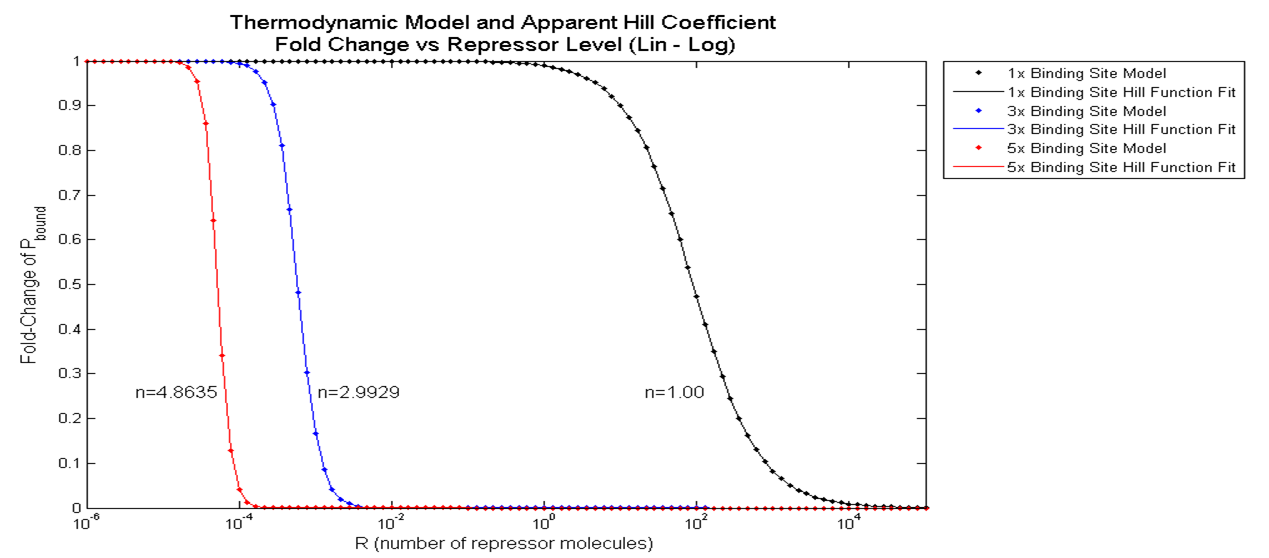
Gene Expression Level vs. Number of Repressor Binding Sites
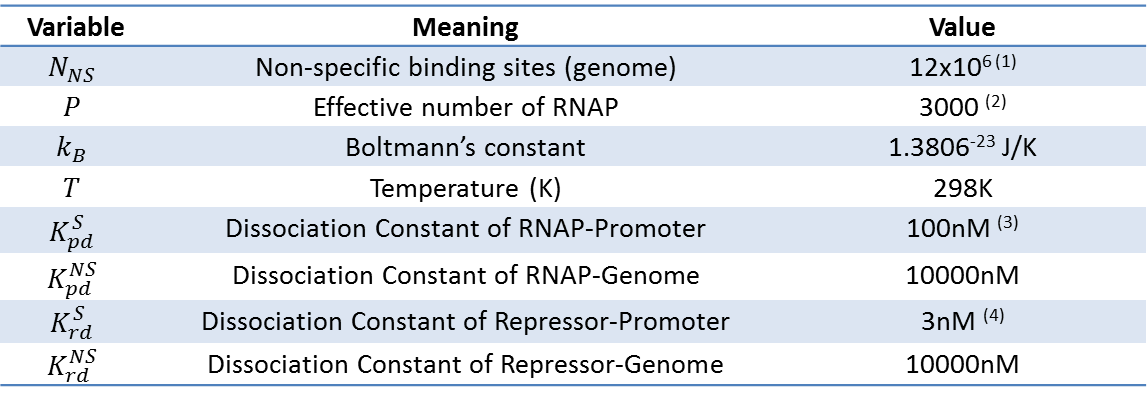
As highlighted in the equation shown above, the relationship between P_bound and the number of F_reg's was studied. First, reasonable values for the variables used were chosen after research. Websites were used to find the size of yeast genome (12e6 bps) and the number of RNAP molecults in a S.cerevisiae (2000~4000 RNAP/cell). Some journal papers were used as well, for example, to find the dissociation constants of a typical TALE (18 RVD's). The variables and their values are summarized below.
Numbers from:
- http://www.yeastgenome.org/cache/genomeSnapshot.html
- http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=109211&ver=3&trm=polymerase
- Bintu et al., "Transcriptional regulation by the numbers". Current Opinion in Genetics and Development. 15, pp. 116-124 (2005)
- Meckler et al., "Quantitative analysis of TALE-DNA interactions suggests polarity effects". Nucl. Acids Res. (2013) 41 (7): 4118-4128.
Gene Expression Level vs. Number of Repressor Binding Sites
 "
"