Team:Duke/Project/Results/Part1
From 2013.igem.org
(Difference between revisions)
(→Results: Library of parts) |
Hyunsoo kim (Talk | contribs) (→Results: Library of DNA Parts for Gene Circuit Construction) |
||
| (35 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | ==Results: Library of | + | {{Template:Team:Duke}} |
| + | <div class="right_box"> | ||
| + | |||
| + | ='''Results: Library of DNA Parts for Gene Circuit Construction'''= | ||
| + | |||
| - | |||
<div>As plasmids were constructed, they were confirmed by restriction digest and sequencing. </div> | <div>As plasmids were constructed, they were confirmed by restriction digest and sequencing. </div> | ||
| + | <br> | ||
| + | |||
| + | ==TALEs== | ||
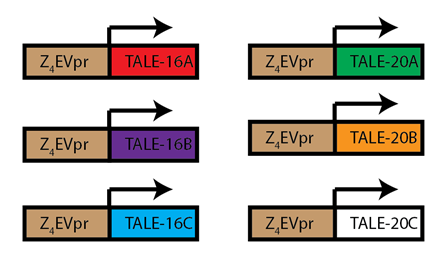
| + | #All 6 TALEs with mCherry tags were built and sequence confirmed | ||
| + | |||
| + | [[File:TaleConstructs.png|300px|center]] <br> | ||
| + | <div align="center"> Figure 1.4.1. TALE Parts Constructed</div> <br> | ||
| + | |||
| + | ==CRISPR== | ||
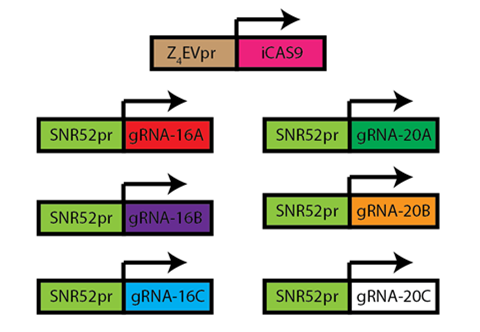
| + | #6 sgRNAs were built and sequence confirmed. | ||
| + | #dCas9-mCherry fusion protein was not cloned successfully, despite many attempts at various cloning protocols. We suspect problems with the source plasmid, or possibly effects of the large insert size. We obtained a dCas9 plasmid without an mCherry fusion that is not yet yeast-integrable. We then cloned a dCas9 plasmid without the fused mCherry gene; this plasmid was sequence confirmed but is yet to be integrated into yeast. | ||
| + | |||
| - | + | [[File:SgRNA_cas9.png|350px|center]] <br> | |
| - | + | <div align="center"> Figure 1.4.2. sgRNA-CRISPR Parts Constructed</div> <br> | |
| - | + | ==Reporters== | |
| - | # | + | #ACT1pr+yEVenus with no operator was built and sequence confirmed |
| - | # | + | #ACT1pr+operator+yEVenus was built and sequence confirmed with 1 and 3 copies of each operator sequence |
| + | #ACT1pr+operator+yEVenus with 5 copies of an operator sequence caused more difficulties, because the operator insert was too repetitive to be synthesized by standard means. We split each sequence into six small oligonucleotides for PCR amplification, but the product was messy and not useful. We then designed two large oligonucleotides for each sequence and were able to PCR amplify and clone the resulting inserts. In this way we obtained and confirmed constructs with 5 copies of certain operator sequences, but not all sequences have been completed. | ||
| + | #After initial testing, we decided to build reporters with TEF1 promoters replacing the ACT1 promoter. TEF1pr+operator+yEVenus with 1 and 3 copies of each operator were built and sequence confirmed. | ||
| - | + | <br> | |
| - | + | [[File:ReporterLibrary.png|370px|center]] <br> | |
| - | + | <div align="center"> Figure 1.4.3. Reporter Parts Constructed</div> <br> | |
| - | + | <br> | |
| - | + | <br><br> | |
| + | </div> | ||
Latest revision as of 23:24, 27 September 2013
Contents |
Results: Library of DNA Parts for Gene Circuit Construction
As plasmids were constructed, they were confirmed by restriction digest and sequencing.
TALEs
- All 6 TALEs with mCherry tags were built and sequence confirmed
Figure 1.4.1. TALE Parts Constructed
CRISPR
- 6 sgRNAs were built and sequence confirmed.
- dCas9-mCherry fusion protein was not cloned successfully, despite many attempts at various cloning protocols. We suspect problems with the source plasmid, or possibly effects of the large insert size. We obtained a dCas9 plasmid without an mCherry fusion that is not yet yeast-integrable. We then cloned a dCas9 plasmid without the fused mCherry gene; this plasmid was sequence confirmed but is yet to be integrated into yeast.
Figure 1.4.2. sgRNA-CRISPR Parts Constructed
Reporters
- ACT1pr+yEVenus with no operator was built and sequence confirmed
- ACT1pr+operator+yEVenus was built and sequence confirmed with 1 and 3 copies of each operator sequence
- ACT1pr+operator+yEVenus with 5 copies of an operator sequence caused more difficulties, because the operator insert was too repetitive to be synthesized by standard means. We split each sequence into six small oligonucleotides for PCR amplification, but the product was messy and not useful. We then designed two large oligonucleotides for each sequence and were able to PCR amplify and clone the resulting inserts. In this way we obtained and confirmed constructs with 5 copies of certain operator sequences, but not all sequences have been completed.
- After initial testing, we decided to build reporters with TEF1 promoters replacing the ACT1 promoter. TEF1pr+operator+yEVenus with 1 and 3 copies of each operator were built and sequence confirmed.
Figure 1.4.3. Reporter Parts Constructed
 "
"