Team:Imperial College/data
From 2013.igem.org
(Difference between revisions)
| Line 10: | Line 10: | ||
<h1>Data</h1> | <h1>Data</h1> | ||
| - | < | + | <h2>Glucose growth assay</h2> |
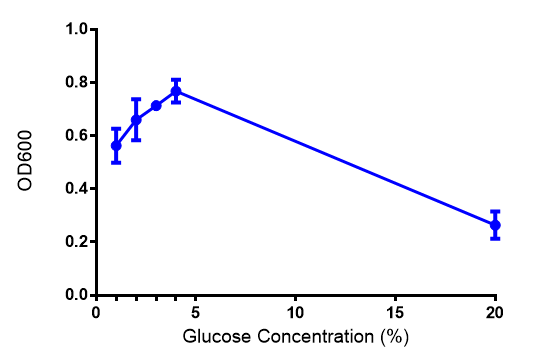
[[File:Glucose_graph.png|thumbnail|center|600px|Cell growth of phaABC <i>E. coli</i> at 4 concentrations of glucose. Optimum growth is at 2-4% glucose at 37ºC. Error bars represents SE of the mean, n=4]] | [[File:Glucose_graph.png|thumbnail|center|600px|Cell growth of phaABC <i>E. coli</i> at 4 concentrations of glucose. Optimum growth is at 2-4% glucose at 37ºC. Error bars represents SE of the mean, n=4]] | ||
<br> <br> | <br> <br> | ||
| - | < | + | <h2>L-lactic Acid growth assay</h2> |
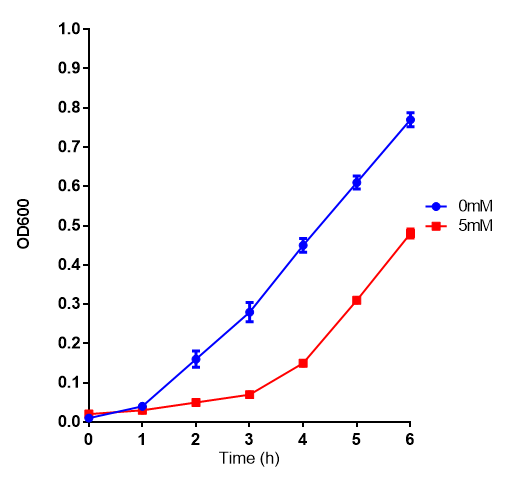
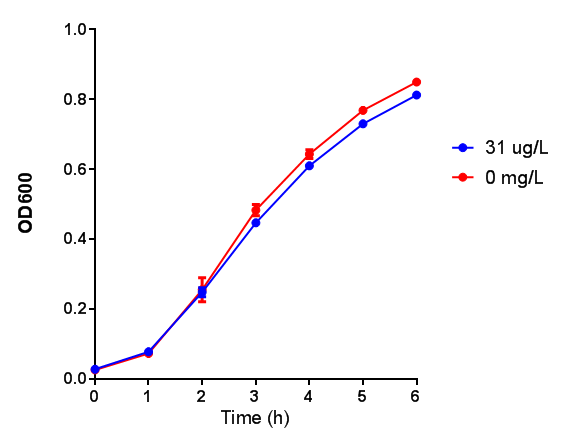
[[File:L-LActic_Acid.png|thumbnail|center|600px|Cell growth of MG1655 on 5mM L-Lactic Acid. Error bars represents SE of the mean, n=4.]] | [[File:L-LActic_Acid.png|thumbnail|center|600px|Cell growth of MG1655 on 5mM L-Lactic Acid. Error bars represents SE of the mean, n=4.]] | ||
<br> <br> | <br> <br> | ||
| Line 37: | Line 37: | ||
<br> <br> | <br> <br> | ||
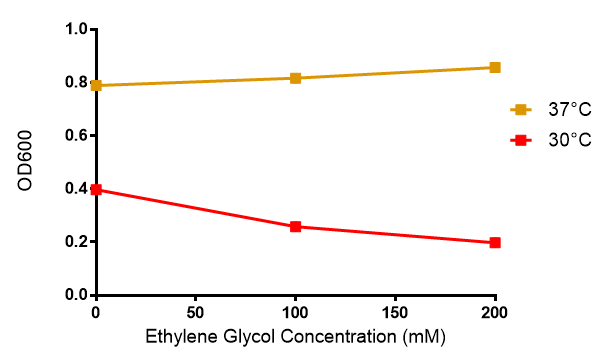
| - | < | + | <h2>Ethylene glycol growth assay</h2> |
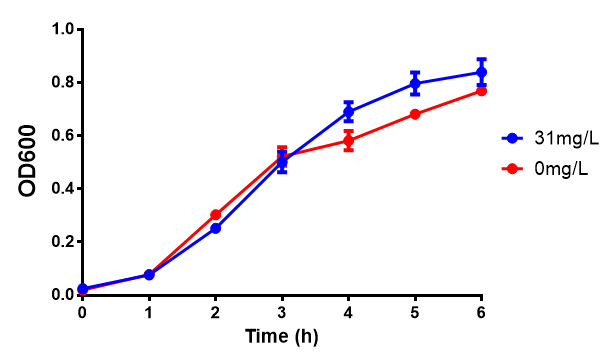
[[File:EG_growth.png|thumbnail|center|600px|Cell growth of MG1655 in ethylene glycol, a byproduct of polyurethane degradation. Cells were grown in 0mM, 100mM or 200mM Ethylene Glycol at 30ºC. Error bars represents SE of the mean, n=4]] | [[File:EG_growth.png|thumbnail|center|600px|Cell growth of MG1655 in ethylene glycol, a byproduct of polyurethane degradation. Cells were grown in 0mM, 100mM or 200mM Ethylene Glycol at 30ºC. Error bars represents SE of the mean, n=4]] | ||
Revision as of 18:28, 21 September 2013
Contents |
BioBricks
<groupparts>iGEM013 Imperial_College</groupparts>
Data
Glucose growth assay
L-lactic Acid growth assay
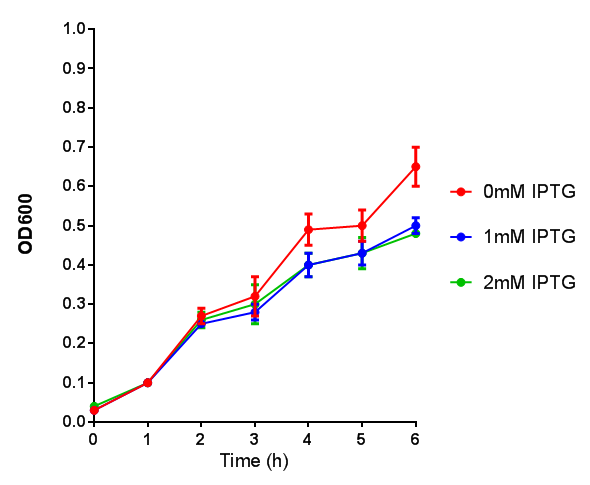
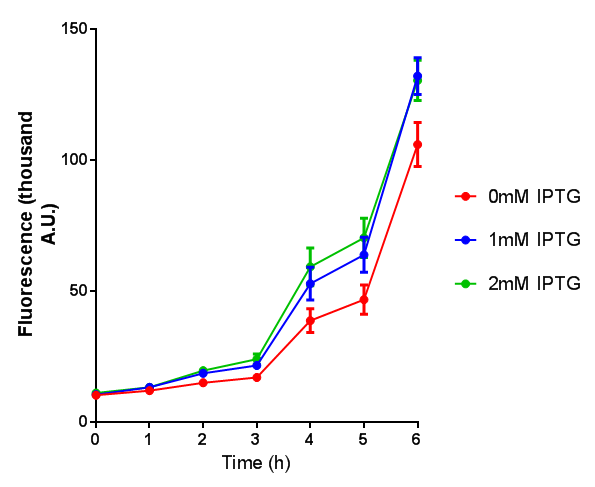
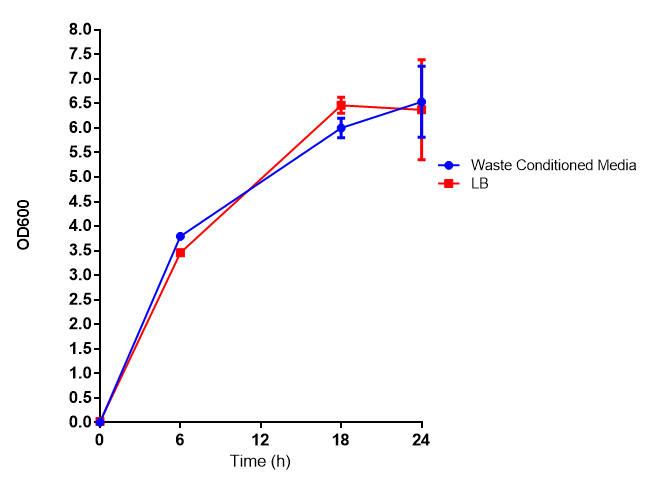
WCM growth and IPTG induction assay
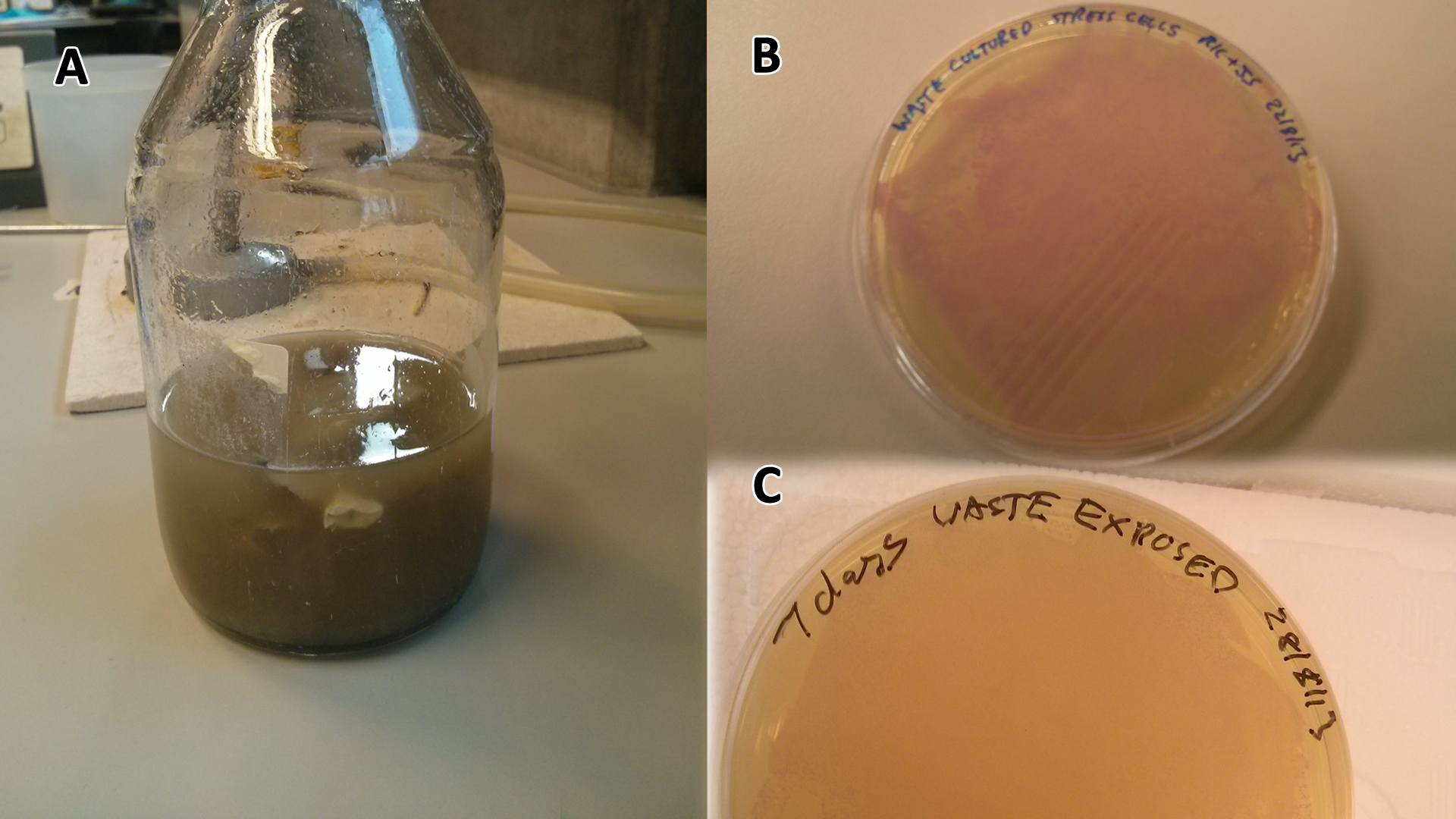
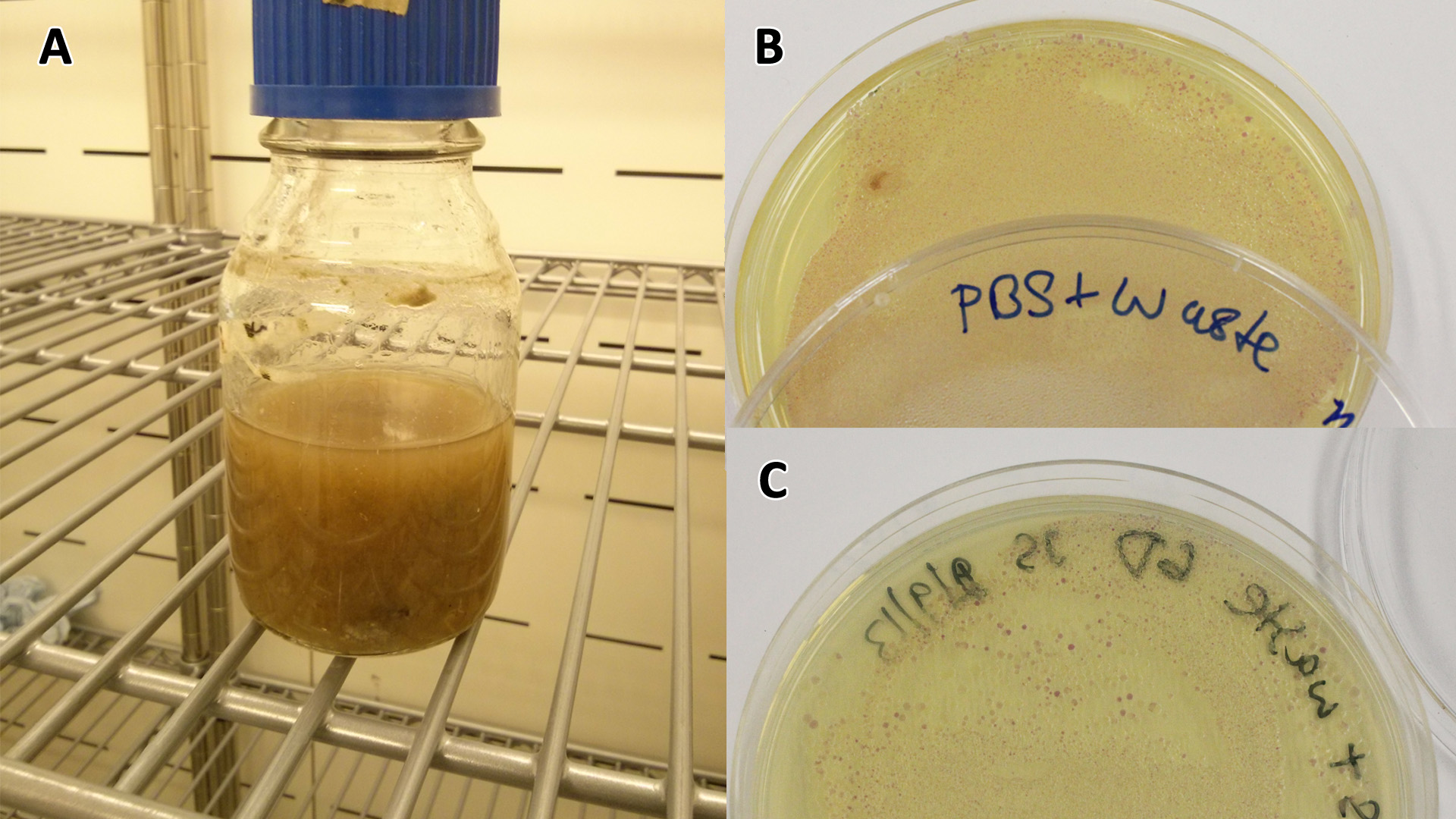
Originally we intended on using [http://parts.igem.org/Part:BBa_K639003 BBa_K639003] to detect whether our cells were stressed when placed in various toxic byproducts. However, as the data below shows, this biobrick is very leaky. As such, we are using the stress sensor as a marker for cell growth and also to show that the cells had been successfully transformed with the correct chloramphenicol resistance.
 Production of the red pigment by stress induction. MG1655 were grown with LB media and sterile filtrated WCM for 48 hours. |
Ethylene glycol growth assay
 "
"