Team:Imperial College/data
From 2013.igem.org
(Difference between revisions)
| Line 4: | Line 4: | ||
<h2>Media characterisation</h2> | <h2>Media characterisation</h2> | ||
| + | <h3>All medias tested</h3> | ||
[[File:LB_M9.png]] | [[File:LB_M9.png]] | ||
| + | <h3>LB</h3> | ||
[[File:LB.png]] | [[File:LB.png]] | ||
| + | <h3>M9 Minimal</h3> | ||
[[File:M9M.png]] | [[File:M9M.png]] | ||
| + | <h3>M9 Supplemented</h3> | ||
[[File:M9S.png]] | [[File:M9S.png]] | ||
| + | <h3>Waste Conditioned Media</h3> | ||
[[File:WCM2.png]] | [[File:WCM2.png]] | ||
| - | + | <h2>Waste Assessment</h2> | |
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
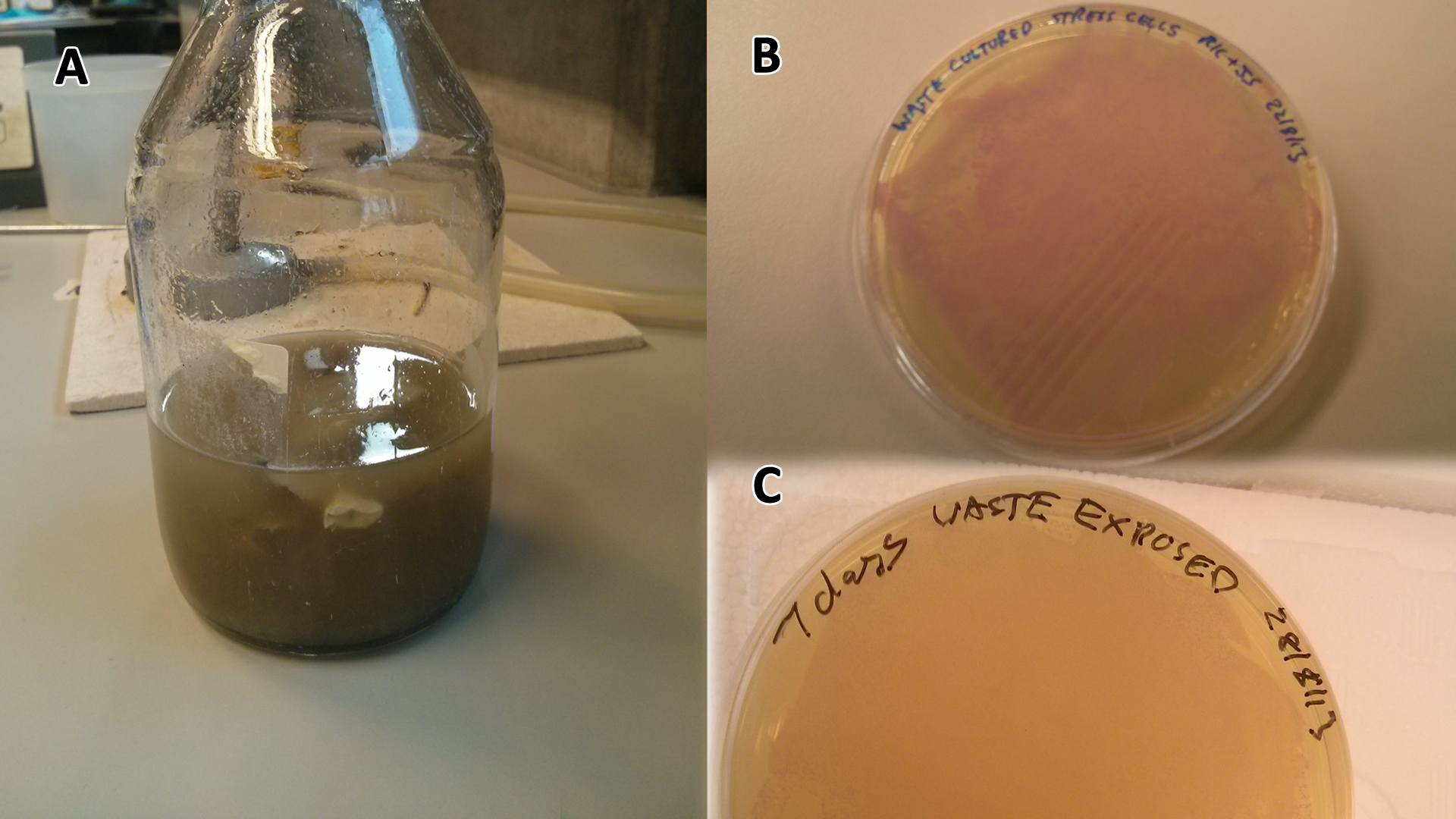
[[File:Waste_cocktail.png|thumbnail|center|600px|<b>(A)</b> WCM precursor material, this sterilised media made from LB and SRF was used to produce all WCM utilised. <b>(B)</b> Cells containing mCherry pigment grown in SRF <b>(A)</b> over 3 days, then streaked in a qualitative assay to check for growth. <b>(C)</b> mCherry cells were streaked again after 7 days growth in SRF.]] | [[File:Waste_cocktail.png|thumbnail|center|600px|<b>(A)</b> WCM precursor material, this sterilised media made from LB and SRF was used to produce all WCM utilised. <b>(B)</b> Cells containing mCherry pigment grown in SRF <b>(A)</b> over 3 days, then streaked in a qualitative assay to check for growth. <b>(C)</b> mCherry cells were streaked again after 7 days growth in SRF.]] | ||
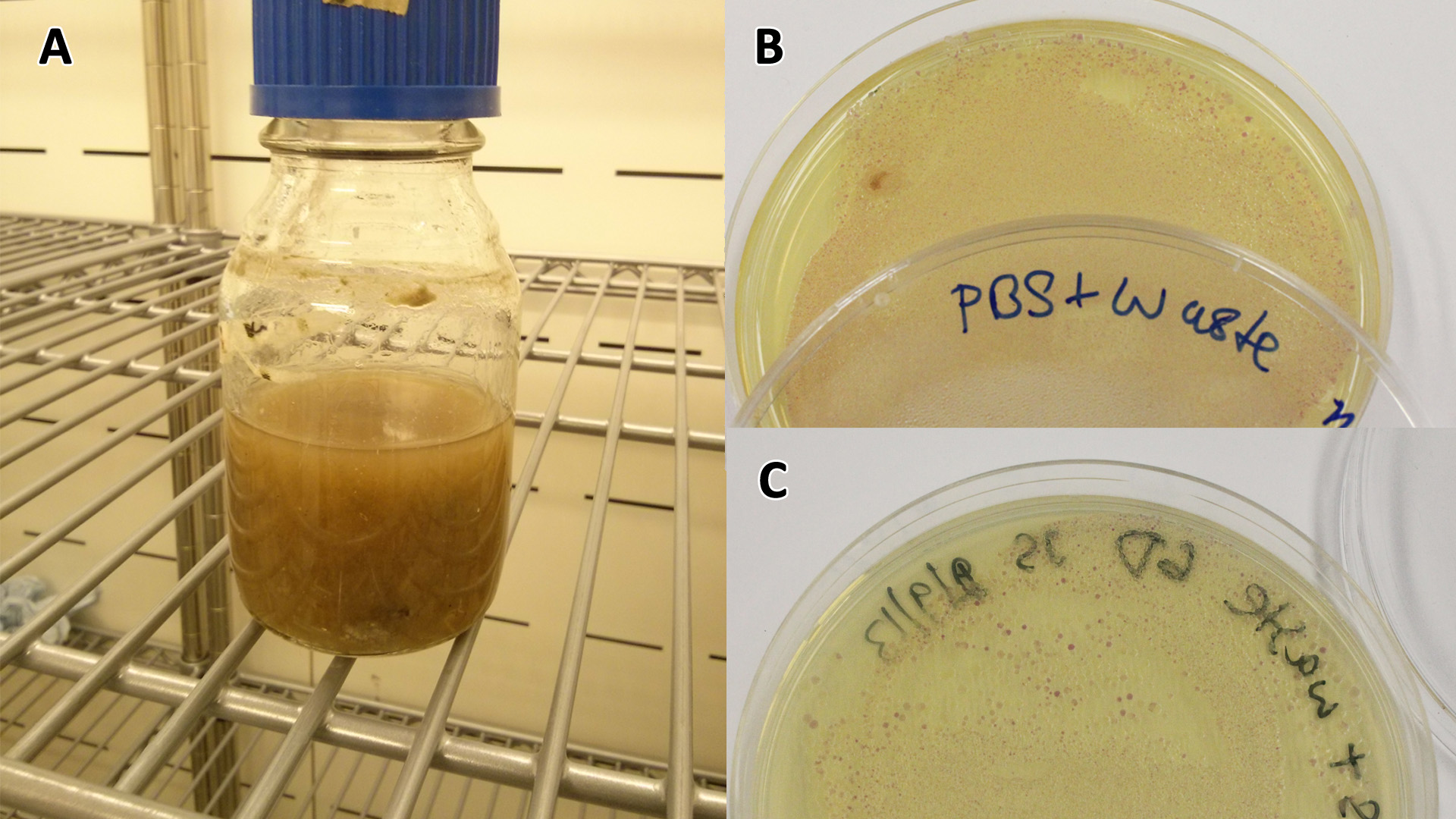
[[File:PBS_Plus_Waste.jpg|thumbnail|center|600px|<b>(A)</b> SRF in PBS (phosphate buffered saline), a buffer. We can see from this experiment whether our bacteria can grow solely on the waste SRF. <b>(B)</b> Cells containing mCherry pigment grown in SRF <b>(A)</b> over 3 days, then streaked in a qualitative assay to check for growth. <b>(C)</b> mCherry cells were streaked again after 6 days growth in SRF.]] | [[File:PBS_Plus_Waste.jpg|thumbnail|center|600px|<b>(A)</b> SRF in PBS (phosphate buffered saline), a buffer. We can see from this experiment whether our bacteria can grow solely on the waste SRF. <b>(B)</b> Cells containing mCherry pigment grown in SRF <b>(A)</b> over 3 days, then streaked in a qualitative assay to check for growth. <b>(C)</b> mCherry cells were streaked again after 6 days growth in SRF.]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
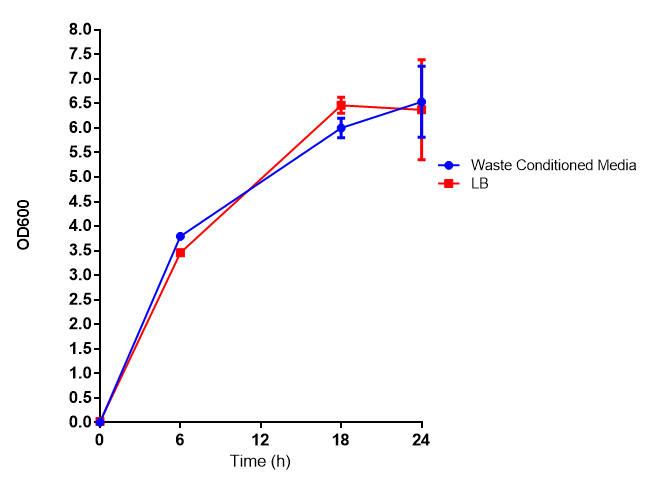
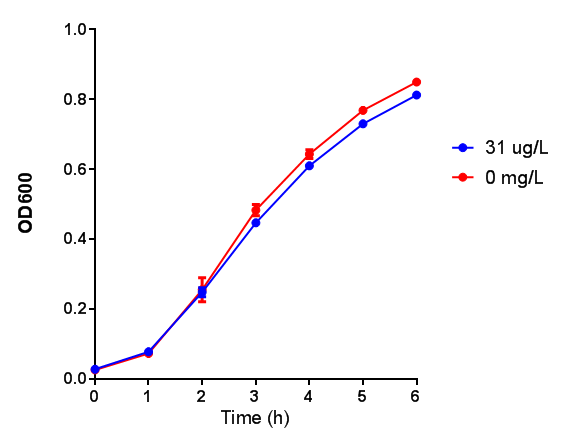
| + | |[[File:WCM_media.png|thumbnail|right|400px|Growth curve of our [http://parts.igem.org/Part:BBa_K639003 mCherry] MG1655 bacteria. MG1655 were grown with LB media and sterile filtrated WCM at 37ºC. Error bars represents SE of the mean, n=4]] | ||
| + | |||
| + | |[[File:Very_stressed_ecoli.jpg|thumbnail|left|400px|Production of the red pigment by stress induction. MG1655 were grown with LB media and sterile filtrated [https://2013.igem.org/Team:Imperial_College/Protocols WCM] for 48 hours.]] | ||
| + | |- | ||
| + | |||
|} | |} | ||
| Line 59: | Line 73: | ||
<br> | <br> | ||
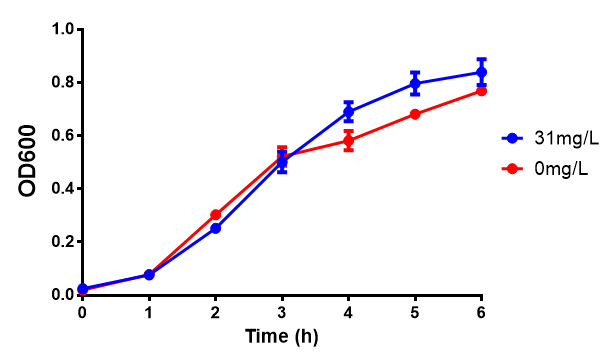
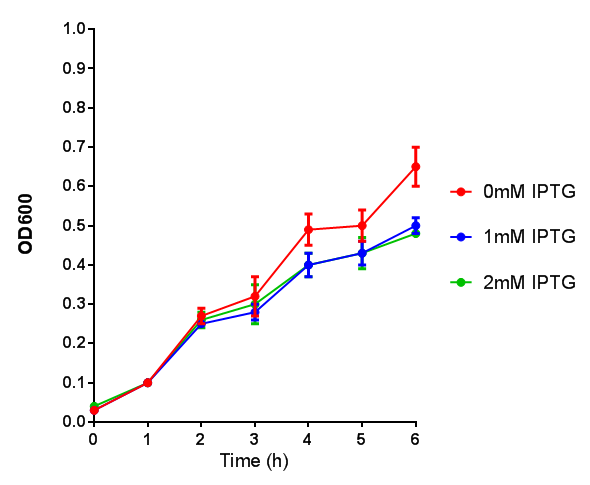
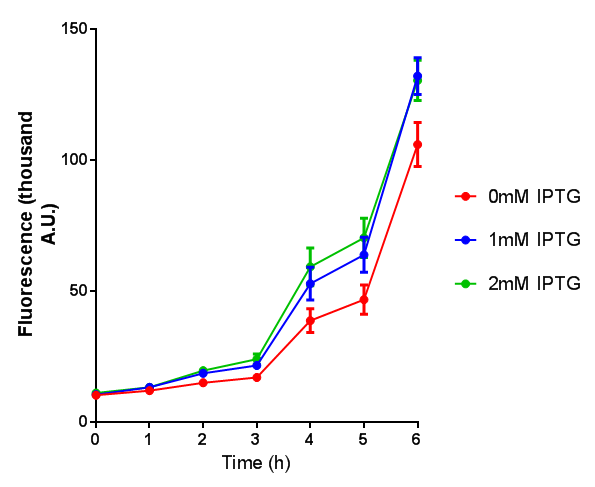
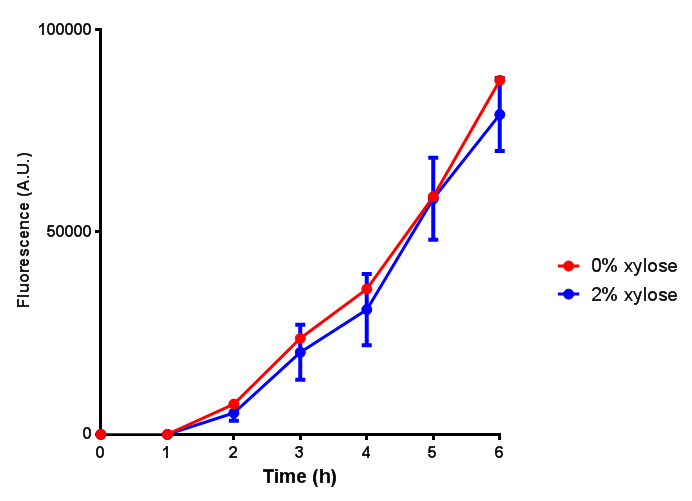
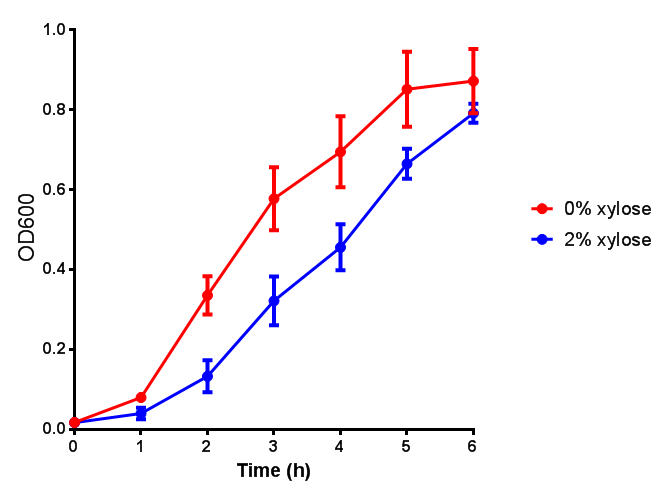
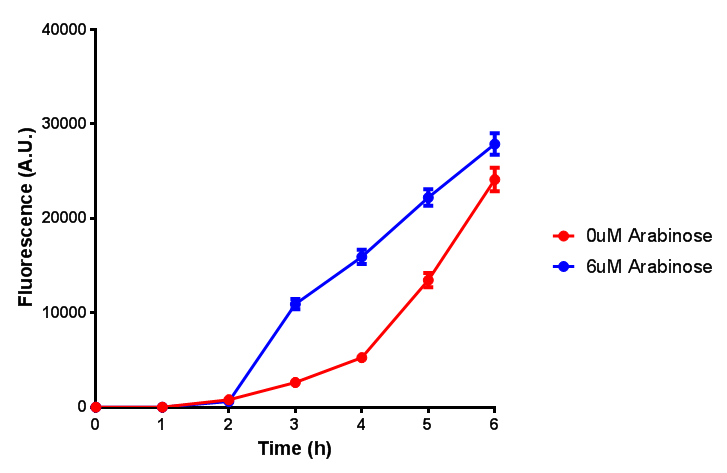
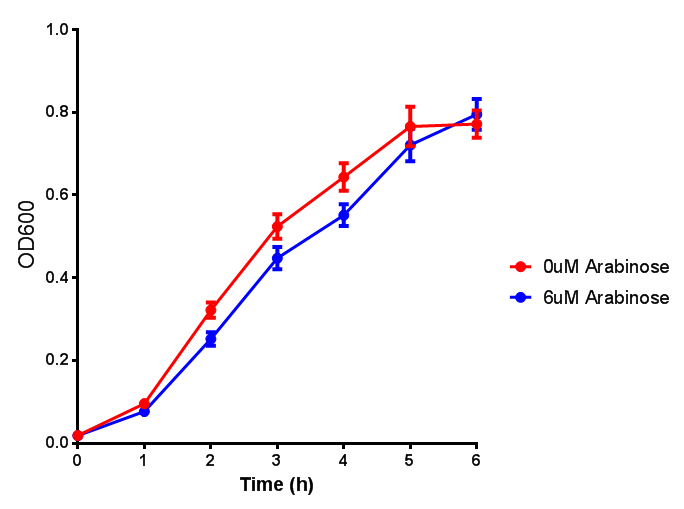
| - | <h3> | + | <h3>IPTG induction assay</h3> |
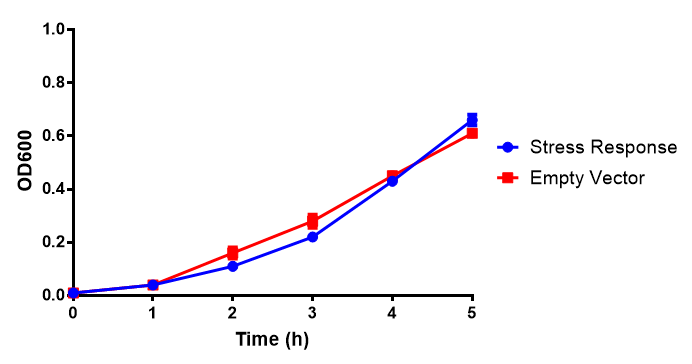
Originally we intended on using [http://parts.igem.org/Part:BBa_K639003 BBa_K639003] to detect whether our cells were stressed when placed in various toxic byproducts. However, as the data below shows, this biobrick is very leaky. As such, we are using the stress sensor as a marker for cell growth and also to show that the cells had been successfully transformed with the correct chloramphenicol resistance. | Originally we intended on using [http://parts.igem.org/Part:BBa_K639003 BBa_K639003] to detect whether our cells were stressed when placed in various toxic byproducts. However, as the data below shows, this biobrick is very leaky. As such, we are using the stress sensor as a marker for cell growth and also to show that the cells had been successfully transformed with the correct chloramphenicol resistance. | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| Line 66: | Line 80: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 16:04, 22 September 2013
Contents |
Growth Assays
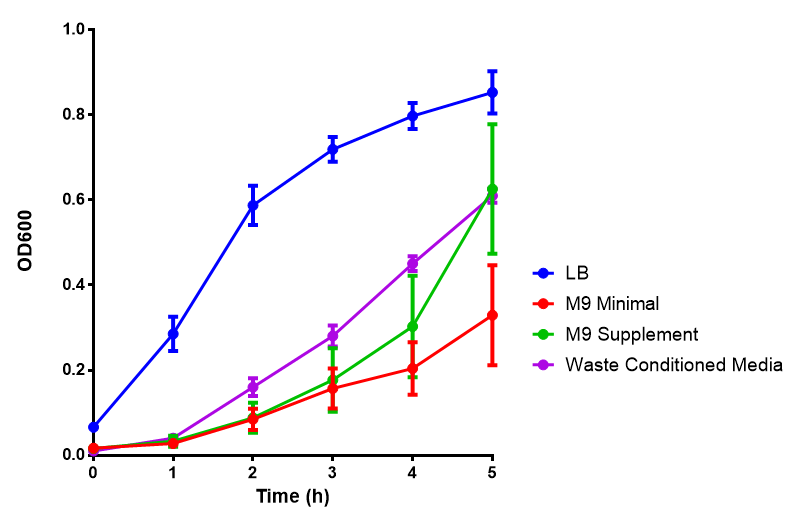
Media characterisation
All medias tested
LB
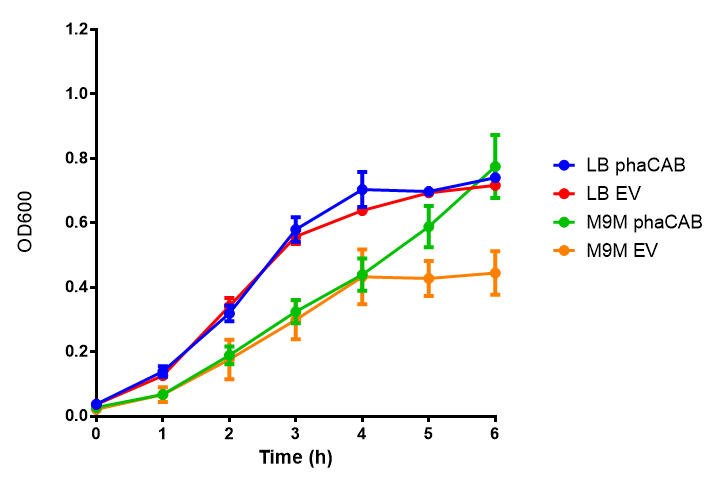
M9 Minimal
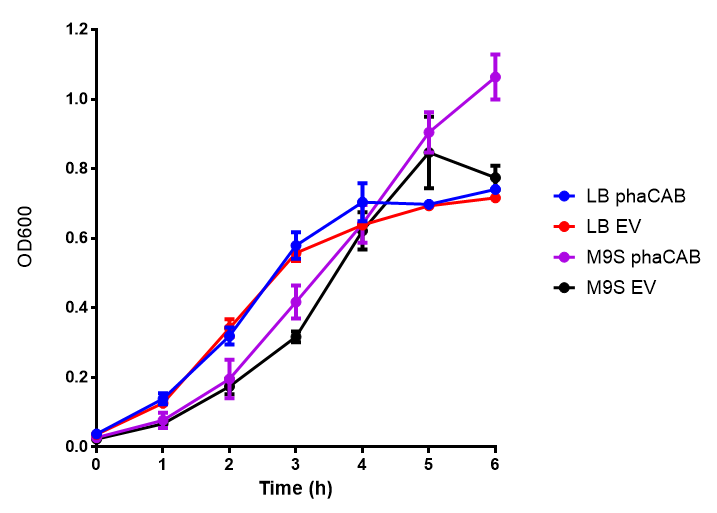
M9 Supplemented
Waste Conditioned Media
Waste Assessment
 Production of the red pigment by stress induction. MG1655 were grown with LB media and sterile filtrated WCM for 48 hours. |
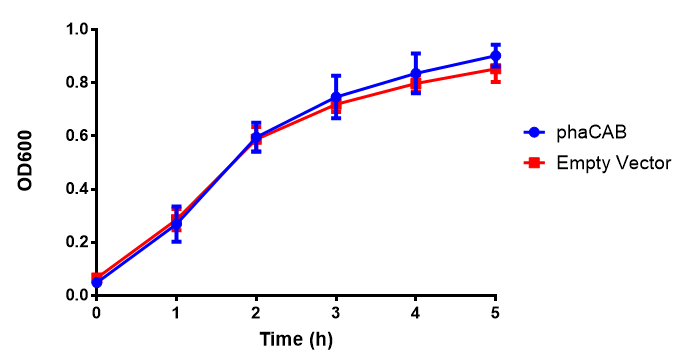
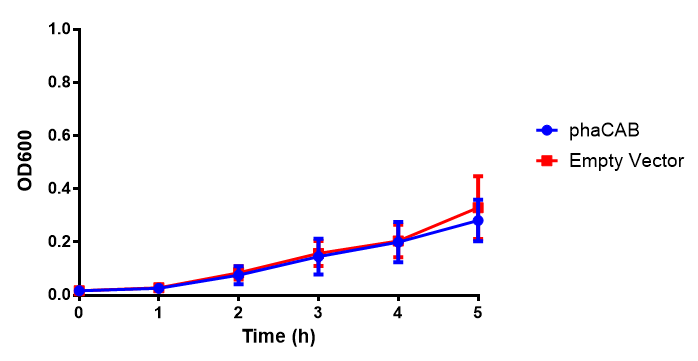
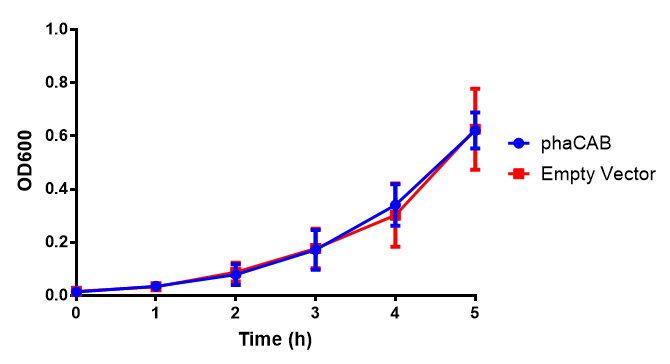
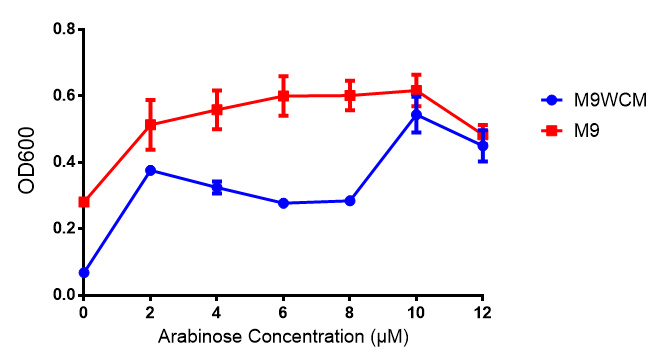
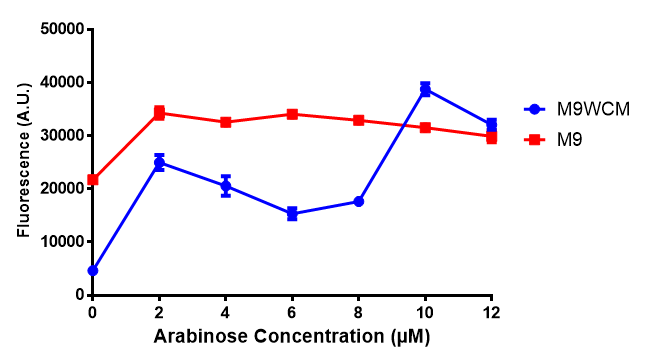
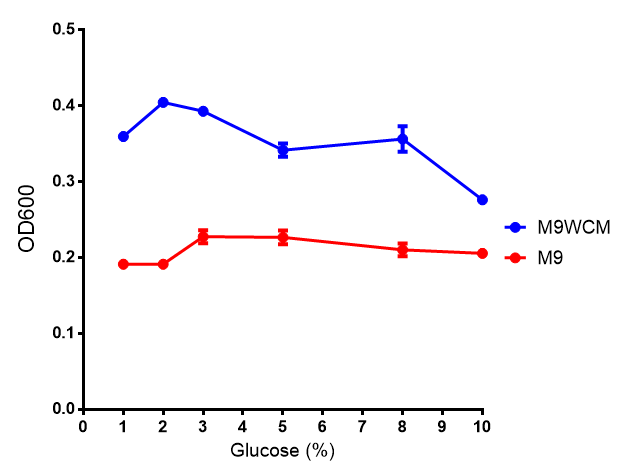
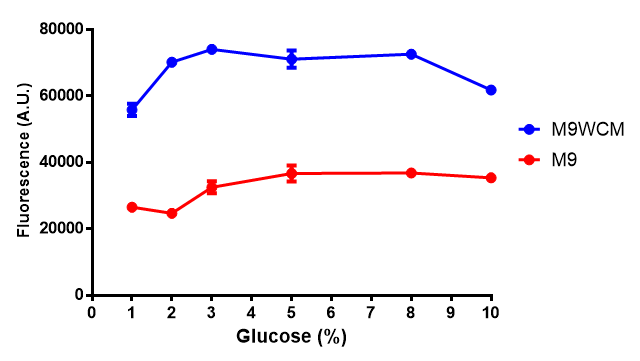
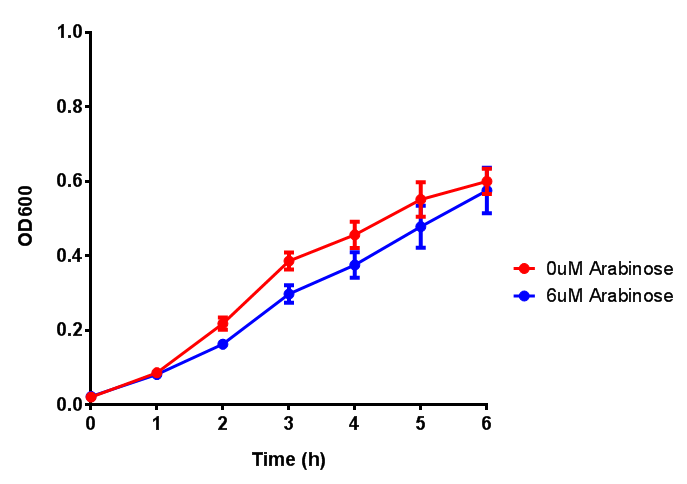
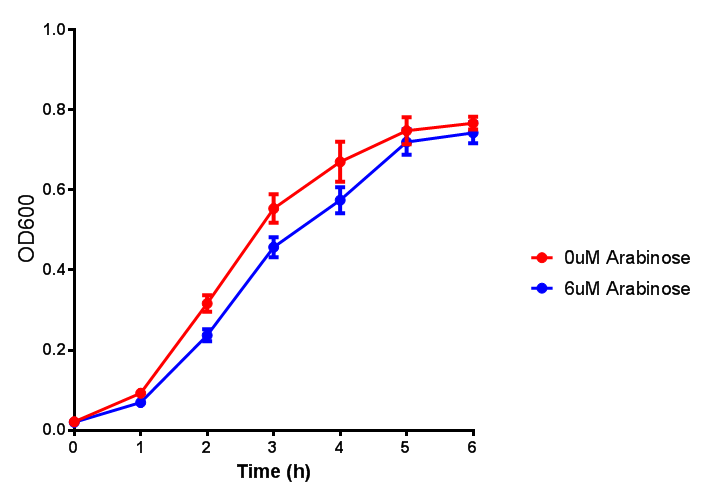
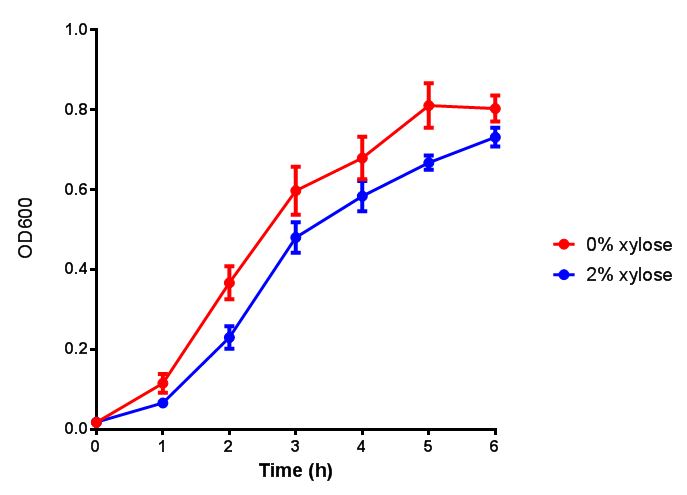
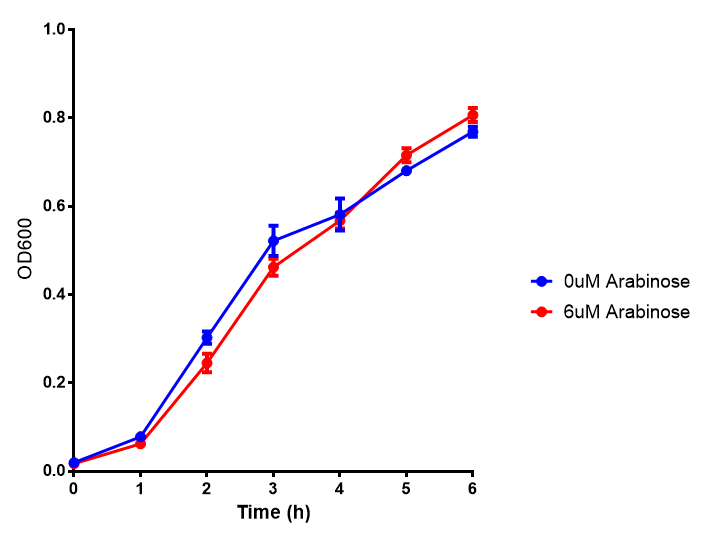
pBAD characterisation
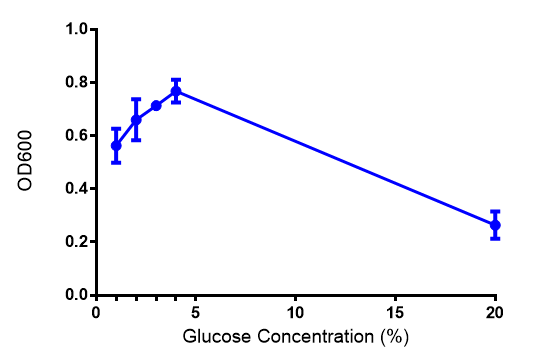
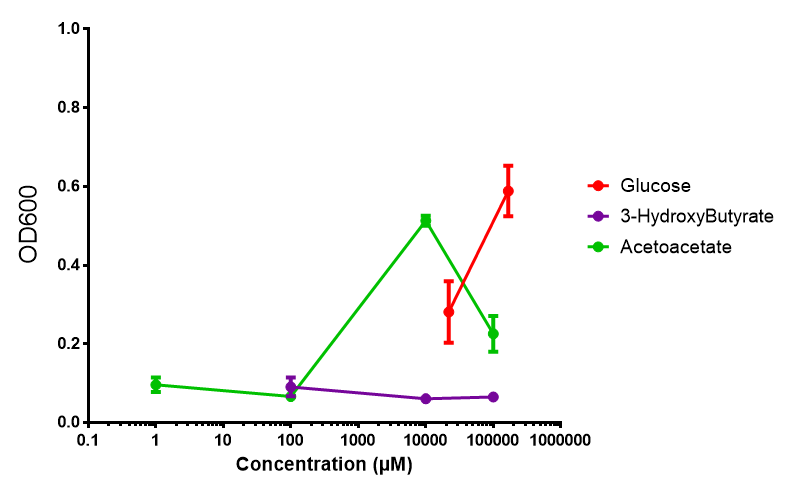
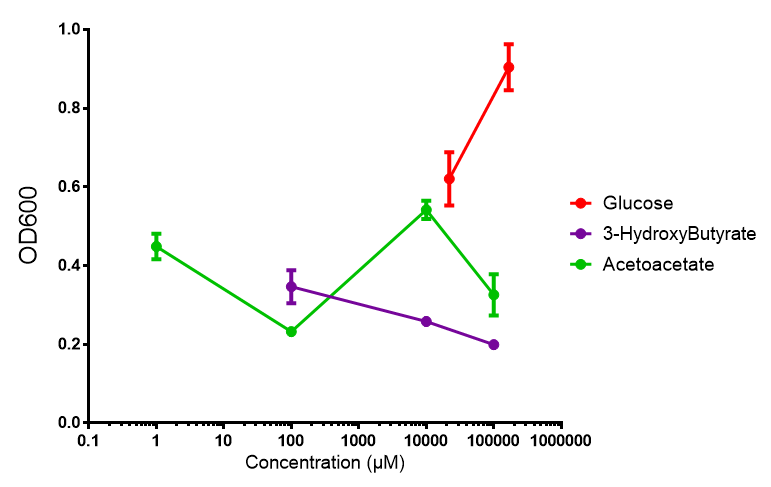
Glucose
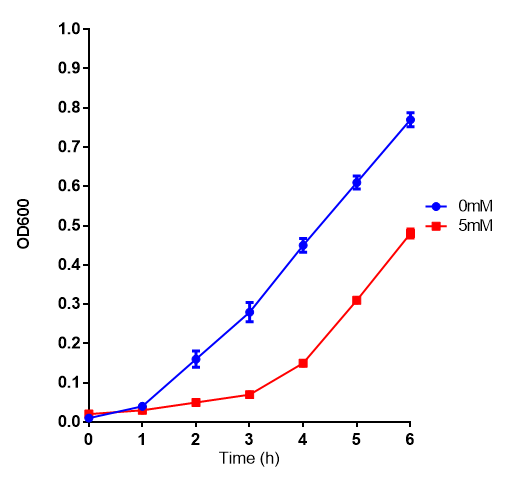
Product inhibition
L-lactic Acid
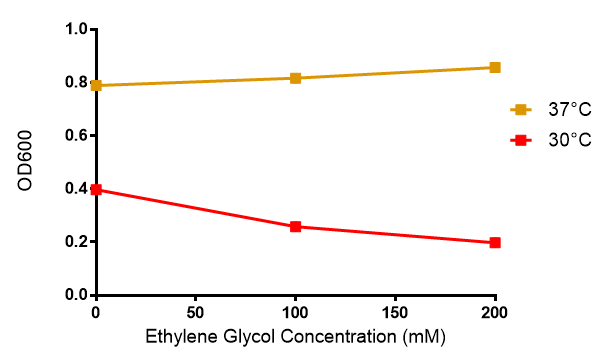
Ethylene glycol
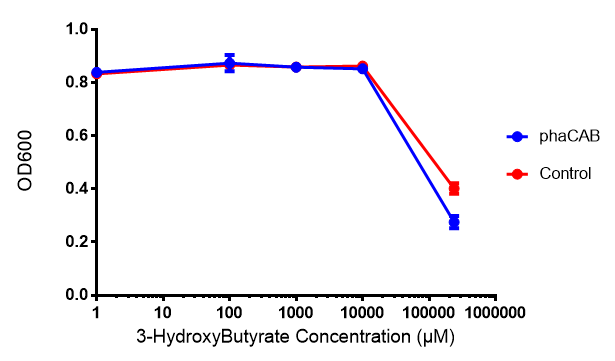
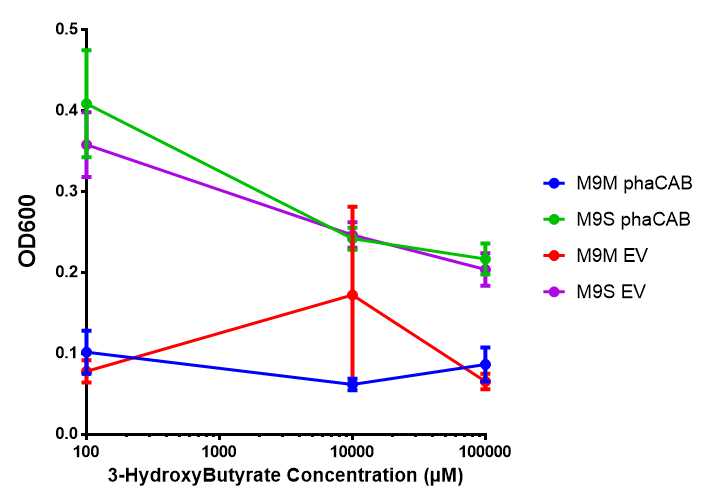
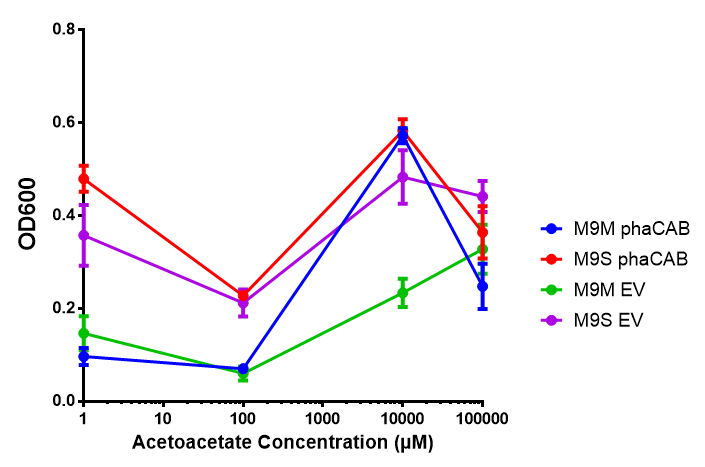
3-hydroxybutyrate (3HB)
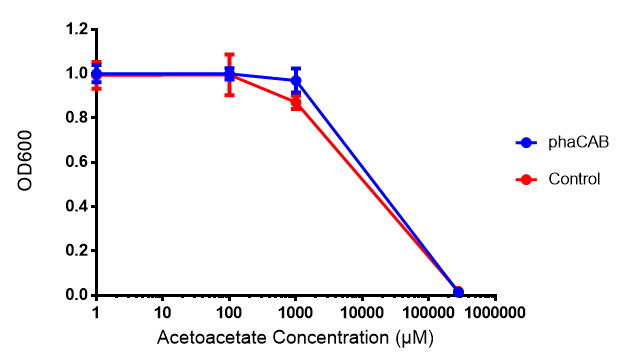
Acetoacetate
Poly(3-hydroxybutyrate) P(3HB)
Poly(lactic acid) (PLA)
IPTG induction assay
Originally we intended on using [http://parts.igem.org/Part:BBa_K639003 BBa_K639003] to detect whether our cells were stressed when placed in various toxic byproducts. However, as the data below shows, this biobrick is very leaky. As such, we are using the stress sensor as a marker for cell growth and also to show that the cells had been successfully transformed with the correct chloramphenicol resistance.
Sole carbon source
3HB
Acetoacetate
Induction
Empty Vector Control
Western blots

Enzyme Kinetics
PHB production
 "
"