Team:NYMU-Taipei/Modeling/Ethanol
From 2013.igem.org


Contents |
Ethanol model
Background:
TrxC promoter is an oxyR-activated promoter. Behind trxC promoter are several terminals to inhibit T7 polymerase-producing gene from being opened easily ; namely, it will not open until the concentration of oxyR is high enough, which means the bees are facing disastrous infection. Once oxyR concentration overcomes the threshold and conquers the terminal obstacles, T7 polymerase is produced and will bind to T7 promoter, which is a specific promoter binding only to T7 polymerase. Behind the T7promoter are enzyme PDC and ADH-producing genes, which will convert pyruvate to ethanol, and thus kill spores of Nosema Ceranae.
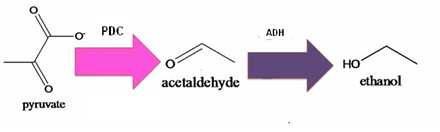
Pathway of ethanol:
PDC= pyruvate decarboxylase; ADH= Acetaldehyde
Aims
1.To simulate how many terminal do we need as a threshold to have the T7 polymerase-producing gene open at a proper time.
2.To simulate the threshold concentration of oxyR to conquer the terminal.
3.To determine the time of opening to see if the circuit could open in time (useful).
As for how we measure promoter strengths, we choose PoPS, which is the rate of RNA polymerase binding to the DNA and trigger the transcription of the certain gene.
It is assumed that the dose of pyruvate is sufficient in bees’ body through enough proliferation/copy number of Beecoli; the possibility of RNApolymerase skipping terminators is assumed to be proportional to time span; equilibrium between pyruvate and acetaldehyde is dominantly rightward, while equilibrium between acetaldehyde and ethanol is bidirectional; concentration of ethanol will not easily decrease and will sustain for a period of time.
Equation1:
1.KdROSoxyR = dissociation constant of ROSoxyR
2.n ROSoxyR = Hill coefficient of ROSoxyR
3.PoPStrxC = promoter strength of trxC
4.KdT7 = dissociation constant of T7
5.NT7 = Hill coefficient of T7
6.PoPST7 = promoter strength of T7 promoter
7.KdegmRNA = degrading constant of sensor promoter mRNA
8.N = number of plasmid in a single cell
9.V = volume of a cell
Equation2:
1.PoPStrxC = promotor strength of trxC promoter
2.PoPST7 = promoter strength of T7 promoter
3.RBS = binding site strength
4.kdegT7 = degrading constant of T7 polymerase
5.a = posibility of conquering the threshold concentration
6.n1 = Hill coefficient of ROSoxyR (complex of ROS+oxyR)
7.N = number of plasmid in a single cell
8.V = volume of a cell
The aim of the equation is to know the threshold concentration of oxyR to conquer the terminal and when T7 polymerase can reach the required concentration to activate T7 promoter.
Equation3:
1.KdT7 = dissociation constant of T7
2.NT7 = Hill coefficient of T7
3.PoPST7 = promoter strength of T7 promoter
4.KdegmRNA = degrading constant of sensor promoter mRNA
5.N = number of plasmid in a single cell
6.V = volume of a cell
The aim of the equation is to know mRNA of enzyme PDC production rate and when it can reach the level to translate enough PDC.
Equation4:
1.RBS = binding site strength
2.PoPST7 = promoter strength of T7 promoter
3.N = number of plasmid in a single cell
4.V = volume of a cell
5.KdegPDC = degrading constant of PDC (pyruvate decarboxylase)
The aim of the equation is to know PDC production rate and when it can reach the concentration of ethanol pathway equilibrium.
Equation5:
1.KdT7 = dissociation constant of T7
2.NT7 = Hill coefficient of T7
3.PoPST7 = promoter strength of T7 promoter
4.KdegmRNA = degrading constant of sensor promoter mRNA
5.N = number of plasmid in a single cell
6.V = volume of a cell
Equation6:
1.RBS = binding site strength
2.PoPST7 = promoter strength of T7 promoter
3.N = number of plasmid in a single cell
4.V = volume of a cell
5.KdegADH = degrading constant of ADH (Acetaldehyde)
The aim of the equation is to know ADH production rate and when it can reach the concentration of ethanol pathway equilibrium.
Equation7:
1.Kpyruvateacetaldehyde = pyruvate→acetaldehyde reaction rate constant
2.Kacetaldehydeethanol = acetaldehyde→ethanol reaction rate constant
3.Km = ethanol→acetaldehyde reaction rate constant
4.KdADH = dissociation constant of ADH
The aim of the equation is to know ethanol production rate and when it can reach the concentration to kill the spores of Nosema Ceranae.
Explanation:
PoPS represents the promoter strength which is measured by the rate of RNApolymerase binding to the starting site of DNA transcription; \frac{ [molecule]^{nmolecule} }{ Kdmolecule^{nmolecule}+[molecule]^{nmolecule} } represents the hill effect of transcription factors (either activators or repressors) to the promoter.
In this equation, the production rate of mRNAT7 is composed of both synthesizing rate and degrading rate. \frac{ [ROSoxyR]^{nROSoxyR} }{ KdROSoxyR^{nROSoxyR}+[ROSoxyR]^{nROSoxyR} }\times{PoPStrxC}\times\frac{N}{V}\times{a} represents how activator ROSoxyR influences promoter trxC on synthesizing mRNAT7 and how thresholds behind the promoter blocks the synthesis of mRNAT7 (the possibility of RNA polymerase to skip thresholds right after trxC promoter is shown in probability);\frac{ [T7]^{nT7} }{ KdT7^{nT7}+[T7]^{nY7} }\times{PoPST7}\times\frac{N}{V}represents how T7 polymerase influences promoter T7 on synthesizing mRNAT7; -KdegmRNA\times[mRNAT7] represents the natural degrading rate of mRNAT7.
In this equation, the production rate of T7 polymerase is composed of both synthesizing rate and degrading rate. RBS represents ribosome binding site strength, which is the affinity of ribosome to the starting site of mRNA; PoPS represents the promoter strength and \frac{[molecule]^{nmolecule}}{ Kdmolecule^{nmolecule}+[molecule]^{nmolecule} }, as mentioned above, represents the hill effect of transcription factors to the promoter.
For the section of the equation,\RBS\times{PoPStrxC}\times\frac{N}{V}\times{a}\times{\frac{ [ROSoxyR]^{n1} }{ [Kd]^{n1}+[ROSoxyR]^{n1} }}} represents the synthesizing rate of T7 polymerase under the influence of activator ROSoxyR complex, promoter trxC and terminators downstream; \PoPST7\times{T7 effect}\times\frac{N}{V}, similarly, represents the synthesizing rate of T7polymerase under the influence of T7promoter; \-{KdegT7}\times{T7}represents the natural degrading rate of T7 polymerase.
In these two equations, the production rate of mRNAPDC and mRNAADH is composed of both synthesizing rate and degrading rate. \frac{ [T7]^{nT7} }{ KdT7^{nT7}+[T7]^{nY7} }\times{PoPST7}\times\frac{N}{V} represents how T7 polymerase influences promoter T7 on synthesizing mRNAPDC and mRNAADH;\frac{ [T7]^{nT7} }{ KdT7^{nT7}+[T7]^{nY7} } and\frac{ [T7]^{nT7} }{ KdT7^{nT7}+[T7]^{nY7} } represents the natural degrading rate of mRNAPDC and [mRNAADH].
Parameters:
| Model | Parameter | Description | Value | Unit | Reference |
|---|---|---|---|---|---|
| Ethanol
| pyruvate | Initial concentration of pyruvate in MG1655 | 1.18 x 2 | g/L | Expression of pyruvate carboxylase enhances succinate production in Escherichia coli without affecting glucose uptake |
| nPDC | Hill coefficient of PDC | 2.1 | - | Purification, characterization and cDNA sequencing of pyruvate decarboxylase Zygosaccharomyces biporus | |
| nADH | Hill coefficient of ADH | at high pH values, 30◦C, n=1; at low temperature, n=3 | - | Evidence for co-operativity in coenzyme binding to tetrameric Sulfolobus
solfataricus alcohol dehydrogenase and its structural basis: fluorescence, kinetic and structural studies of the wild-type enzyme and non-co-operative N249Y mutant |
 "
"