Team:Paris Saclay/Modeling
From 2013.igem.org
(→Complex simulation) |
(→Simple simulation) |
||
| Line 141: | Line 141: | ||
=== Simple simulation === | === Simple simulation === | ||
| - | {{Team:Paris_Saclay/simbox | + | {{Team:Paris_Saclay/simbox|load=modeling/fnr0.xml}} |
=== Complex simulation === | === Complex simulation === | ||
Revision as of 20:45, 27 September 2013
Contents |
Modeling
"La modélisation, c'est comme le sexe, plus y'a d'inconnues, plus c'est chaud." ---damdam_du_91
Before you start, maybe those words could make you understand more quickly our project.
What do we model?
This modeling is about the simulation of FNR aero/anaerobic regulation system which we have successfully achieved in our experiments. The second part of our experiment: the PCBs sensor system is not touched in the modeling.
The FNR aero/anaerobic regulation system, briefly, we try to rebuild mathematically a system which can simulate the alternate expression of FNR in aerobic or anaerobic situation, from the translation of fnr to the production of reporter protein.
What’s our purpose for this modeling?
The main aim of our modeling is recreate the aero/anaerobic reporter protein expression system. By alternating the oxygen concentration, activated or inactivated FNR combine (or not) with potential sensor gene sequence and make blue or red color. Our job is to see if we can observe the alternating red and blue curve when we change the oxygen condition.
How do we model?
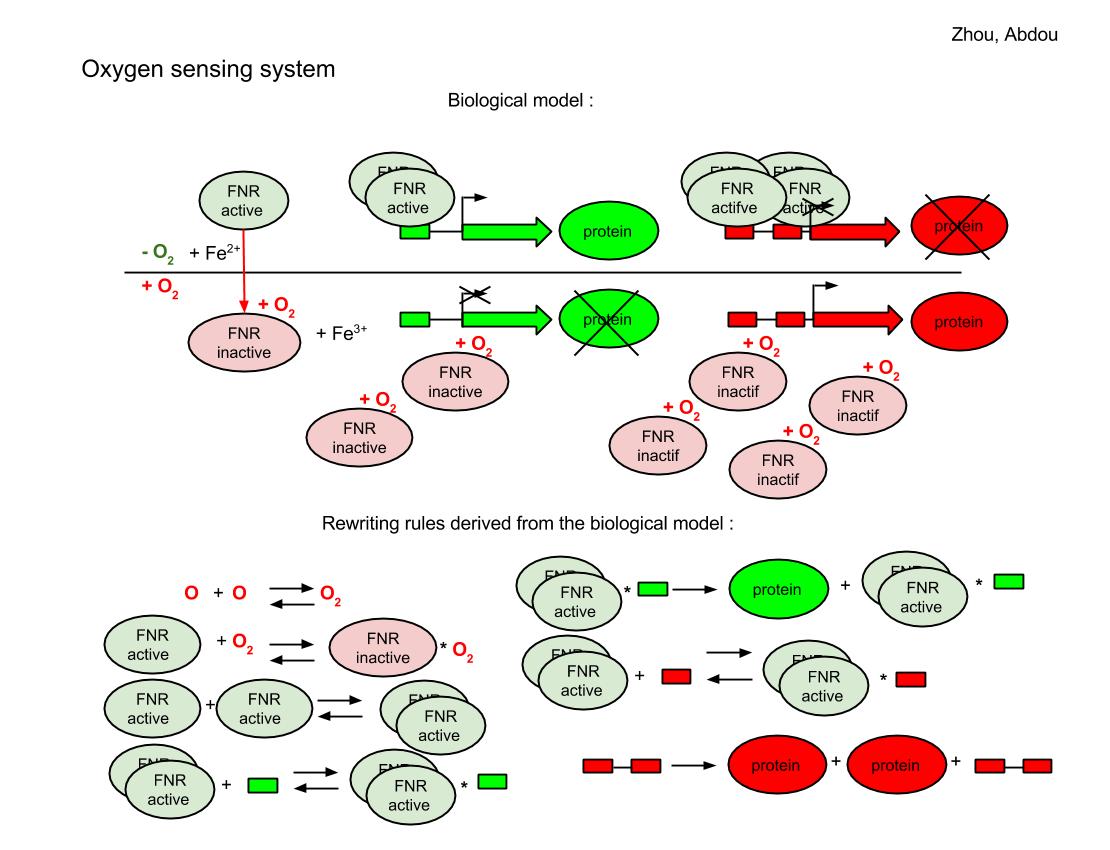
As the research of FNR regulation system is a well-studied territory, we found some excellent scientific paper in library especially [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2867928/ Regulation of Aerobic to Anaerobic Transitions by the FNR cycle in Escherichia coli by Dean A.Tolla and Michael A.Savageau]. This is a differential equation model. And in our modeling we decided to redo what they have done but use a different method: a stochastic system created by new modeling tool: Hsim. Hsim is a stochastic automation simulator, it simulate chemical or enzymatic reactions between molecules and based on reactions it can simulate whole system.
Oxygen censor, FNR regulator system modeling
As we use Hsim for the modeling, if you want to dig in our model we suggest you to read this page written by Mr. Patrick Amar the developer of Hsim.
Roughly speaking, we define a sampling volume, size in nanometer and we declare some molecules with their sizes, their moving speed and their number. Once the simulation starts, those molecules move randomly in the space and trigger some possible reaction between them with some probabilities. There is always somehow deviation from the reality, but we are all excited about this particular simulating modeling.
We represent our FNR regulator model to you by 3 steps. We have chosen HSIM, a multi-agents programme developped at our university in order to follow the assembly, movements and dissociation of a large number of molecules in a virtual cell. HSIM replaces the Ordinary Differential Equations and mimics chemical reactions of the real system by rewriting rules. Following our modeling process, we provided the list of molecules, the rewriting rules, and the initial values of each molecule quantity in a configuration file to HSIM. We also have developed a web version of Hsim which allows user change and test the system parameters.
Steps of modeling
Step 1: translate biological knowledges into functional scheme
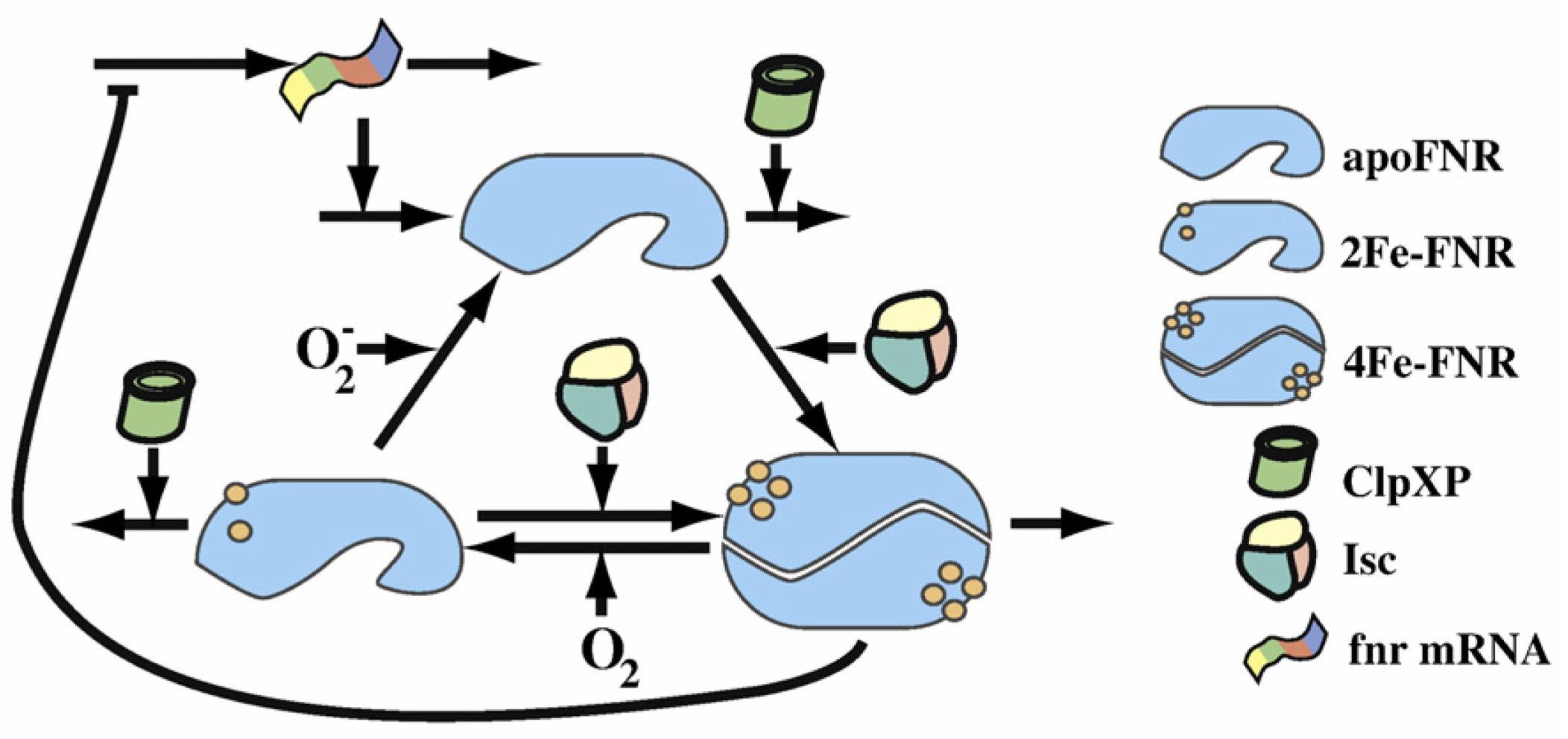
This scheme from the Dean A.Tolla and Michael A.Savageauthe's paper shows clearly the FNR transformation cycle. After the fnr mRNA translation, apoFNR is produced and a dimerization of 2 apoFNR catalyzed by ClpXP forme create the 4Fe-FNR. The 4Fe-FNR can be oxidized by oxygen to 2Fe-FNR then to apoFNR. 2Fe-FNR plus ClpXP can generate 4Fe-FNR. And all 3 FNR derivatives sustain a degradation effect. Those proteins will dissimilate in a while.
If we combine the FNR cycle and color reporter system :
The FNR behave differently for aerobic and anaerobic condition. In the case that oxygen is absent, a pair of 4Fe-FNR fix on the reporter DNA sequence and the blue reporter gene is expressed so we will see the blue color for those colonies. But a group of 4 4Fe-FNR come to fix at red reporter’s gen and prevent the expression of red color. If oxygen is sufficient, FNR derivatives keep in inactive form, the red reporter’s gene continue to produce the red color.
Step 2: transcribe functional description into equation system
Summary for equations:
- translation of Pfnr gene into FNR protein (FNR without Fe)
- dimerisation of 4Fe-FNR (action of Ise protein)
- oxydation: in aerobic conditions, oxygen inactivates FNR but cell continues to reactivate it (action of Ise protein)
- activation/inactivation of a gene under a repressor promotor (green) and production or not of the cognate protein
- activation/inactivation of a gene under an activator promotor (red) and production or not of the cognate protein
Details
- Translating fnr to FNR
- fnr mRNA generates steadily apoFNR(inactived).
- fnr -> fnr + apoFNR [probability]
- Dimerization of apoFNR
- 2 apoFNRs form 4Fe-FNR(activated) with catalyzer Ise.
- {apoFNR}apoFNR + Ise -> QFeFNRa + Ise[probability]
- Oxidation, from 4Fe-FNR to apoFNR(inactivated)
- Aerobically, oxygen inactivates FNR, from 4Fe-FNR to 2Fe-FNR(inactivated)
- QFeFNRa + o2 -> DFeFNRi +o2 [probability]
- Then, oxidize from 2Fe-FNR to apoFNR
- DFeFNRi + o2 -> apoFNR +o2 [probability]
- 2Fe-FNR can be reactivate by Ise
- DFeFNRi + Ise -> QFeFNRa + Ise [probability]
- Degradation FNR
- Protein FNR and its derivation submit a steady degrading rate
- apoFNR + ClpXP -> ClpXP [probability]
- DFeFNRi + ClpXP -> ClpXP [probability]
- QFeFNRa + ClpXP -> ClpXP [probability]
- Activation/inactivation of green gene
- Association between 4Fe-FNR(activated) and green protein gene
- QFeFNRa + GPV -> QFeFNRa_GPV_binded [probability]
- Dissociation between 4Fe-FNR(activated) and green protein gene
- QFeFNRa_GPV_binded -> QFeFNRa + GPV [probability]
- QFeFNRa_GPV_binded -> PV + QFeFNRa_GPV_binded [probability]
- Green protein degradation :catalyzed by ClpXP
- PV + ClpXP -> ClpXP [probability]
- Repression of red protein gene : Association between 4Fe-FNR(activated) and red protein gene
- QFeFNRa + GPR -> QFeFNRa_GPR_binded [probability]
- Dissociation between 4Fe-FNR(activated) and red protein gene
- QFeFNRa_GPR_binded -> QFeFNRa + GPR [probability]
- Association of 4 4Fe-FNR(activated)
- {QFeFNRa_GPR_binded}QFeFNRa_GPR_binded -> rep [probability]
- Dissociation of 4 4Fe-FNR(activated)
- rep -> QFeFNRa_GPR_binded + QFeFNRa_GPR_binded [probability]
- Expression the red gene in aerobic condition
- Production of red protein without repression
- GPR -> GPR + PR [probability]
- QFeFNRa_GPR_binded -> QFeFNRa_GPR_binded + PR [probability]
- Degradation of red protein
- PR + ClpXP -> ClpXP [probability]
Here you will find our configuration files to launch HSIM: links.
3rd step: HSim on a web simulator
Damir has conceived and implemented the web simulations
We provide 2 web versions of the HSIM simulator. Each of them implements a different set of rules. The "Simple simulation" describes the following reactions:
- translation of Pfnr gene into FNR protein (FNR without Fe)
- dimerisation of 4Fe-FNR (action of Ise protein)
- oxydation: in aerobic conditions, oxygen inactivates FNR but cell continues to reactivate it (action of Ise protein)
- degradation of each kind of FNR (FNR without Fe, FNR activated, and FRN inactivated), a green protein, and a red protein (action of ClpXP
protein)
- activation/inactivation of a gene under a repressor promotor (green) and production or not of the cognate protein
- activation/inactivation of a gene under an activator promotor (red) and production or not of the cognate protein
We used the simple simulation to explore different combinations of initial values. The complex simulation itemizes more rules than the simple simulation. en cours
Here you will find our configuration files to launch HSIM: links.
==How to launch the simulations?== When simulation is launched, curve of molecule quantities appear with time. The "START/PAUSE" and "RESET" buttons give control to the simulation time. You may change the oxygen quantities with the "O2" panel.
Oxygen sensing system
Simple simulation
Complex simulation
Results and discussion
en cours
Writing by Zhou and Damir
 "
"