Team:Paris Saclay/Notebook/August/27
From 2013.igem.org
(→summary) |
(→2 - Electrophoresis of the colony PCR products : Pndh* or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in pSB1C3) |
||
| (34 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Paris_Saclay/incl_debut_generique}} | {{Team:Paris_Saclay/incl_debut_generique}} | ||
| + | |||
='''Notebook : August 27'''= | ='''Notebook : August 27'''= | ||
| - | ==''' | + | =='''Lab work'''== |
| - | + | ==='''A - Aerobic/Anaerobic regulation system'''=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==''' | + | ===='''Objective : characterize BBa_K1155000 and BBa_K1155004'''==== |
| - | + | ===='''1 - PCR Colony of ligation Pndh* or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in pSB1C3'''==== | |
| - | + | ||
| - | + | ||
| - | + | XiaoJing | |
| - | + | {| | |
| - | + | | style="border:1px solid black;padding:5px;background-color:#DE;" | | |
| - | + | Transformation of 08/26/13 works. We will do a Colony PCR. | |
| - | + | |} | |
| - | + | ||
| - | + | We took a single colony and resuspend in 10µL H2O.For each Biobrick we did 6 PCR Colony. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Used quantities : | |
| - | + | * DNA : 2µL | |
| + | * Mix : (it was divided in 6 tubes for each promotor with 23µL of mix in each tube) | ||
| + | ** Oligo 44 : 17.5µL | ||
| + | ** Oligo 43 : 17.5µL | ||
| + | ** Buffer Dream Taq : 87.5µL | ||
| + | ** dNTP : 17.5µL | ||
| + | ** Dream Taq : 7µL | ||
| + | ** H2O : 591µL | ||
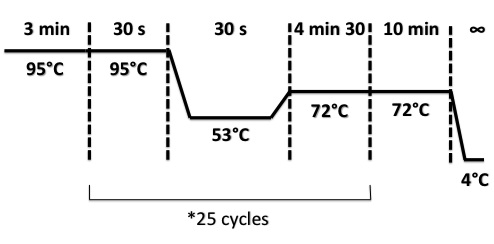
| - | + | PCR Program : | |
| + | [[File:PsPCR2708.jpg|400px]] | ||
| + | ====2 - Electrophoresis of the colony PCR products : Pndh* or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in pSB1C3==== | ||
| + | XiaoJing | ||
| + | {| | ||
| + | | style="width:350px;border:1px solid black;" | [[File:Psgel12708.jpg|500px]] | ||
| + | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
| + | * Well 1 : 6µL DNA ladder | ||
| + | * Well 2 to 7 : 10µL of NirB with RBS-LacZ-Term in pSB1C3 +2µL of 6X loading dye | ||
| + | * Well 8 to 13 : 10µL of NirB with RBS-Amil CP-Term in pSB1C3 +2µL of 6X loading dye | ||
| + | * Well 14 to 19 : 10µL of Pndh* with RBS-LacZ-Term in pSB1C3 +2µL of 6X loading dye | ||
| + | * Well 20 to 25 : 10µL of Pndh* with RBS-Amil CP-Term in pSB1C3 +2µL of 6X loading dye | ||
| + | * Gel : 1% | ||
| + | |} | ||
| + | Expected size : | ||
| + | * NirB with RBS-LacZ-Term in pSB1C3 : 3474bp | ||
| + | * NirB with RBS-AmilCP-Term in pSB1C3 : 1029bp | ||
| + | * Pndh* with RBS-LacZ-Term in pSB1C3 : 3380bp | ||
| + | * Pndh* with RBS-AmilCP-Term in pSB1C3 : 935bp | ||
| - | ===== | + | {| |
| + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| + | We obtained fragments at the right size for NirB with RBS-Amil CP-Term in pSB1C3 in well 12, Pndh* with RBS-LacZ-Term in pSB1C3 in well 14, 15, 18 and 19 and Pndh* with RBS-Amil CP-Term in pSB1C3 in well 20, 22 and 25. Nevertheless, electrophoresis shows that these colonies weren't pure. We will purify them by streaking. | ||
| + | |} | ||
| + | |||
| + | ====3 - Streak colonies of NirB with RBS-Amil CP-Term in pSB1C3, Pfnr with RBS-LacZ-Term in pSB1C3 and Pfnr with RBS-Amil CP-Term in pSB1C3 to purify them==== | ||
| + | |||
| + | XiaoJing | ||
| + | |||
| + | ====4 - Culture of strain MG1655Z1 Δfnr::Km containing plasmid pcp20==== | ||
| + | |||
| + | XiaoJing | ||
{| | {| | ||
| - | | style=" | + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | |
| - | + | Purification from 08/26/13 works. | |
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| - | + | We made culture on LB medium at 42°C for strain MG1655Z1 Δfnr::Km containing plasmid pcp20 to select the bacteria that eliminate the Km cassette. Therefore, we can use this strain to receive the plasmid IGEM pSB1K3. Like this we will select clone Δfnr::Km by streaking them on plate with ampicilin, kanamycin and chloramphenicol plates. | |
| - | + | Principle : | |
| - | + | ||
| + | Plasmid pcp20 is thermosensitive plasmid which produces a flipase that is able to eliminate antibiotic gene (in our case is Km) at 30°C. | ||
| + | When we shift back the strain at 42°C, the plasmid can not replicate and the bacteria will loose their plasmid. | ||
| + | As a result, we constructed successfully the mutant strain MG1655Z1::Δfnr without any antibiotic resistance. | ||
| + | ==='''A - Aerobic/Anaerobic regulation system / B - PCB sensor system'''=== | ||
| + | ===='''Objective : obtaining FNR, BphR2 proteins'''==== | ||
| + | |||
| + | ===='''1 - Electrophoresis of RBS-BphR2 Part I, BphR2 Part II, FNR Part I , FNR Part II, RBS-FNR Part I and pSB1C3'''==== | ||
| + | |||
| + | XiaoJing | ||
{| | {| | ||
| - | | style=" | + | | style="border:1px solid black;padding:5px;background-color:#DE;" | |
| - | + | Gibson tranformation of the 08/27/13 didn't work. So we did an electrophoresis to check sizes and concentrations of Gibson parts. | |
| - | + | |} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | *Gel : 1 | + | {| |
| + | | style="width:350px;border:1px solid black;" | [[File:Psgel22708.jpg]] | ||
| + | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
| + | * Well 1 : 6µL DNA ladder | ||
| + | * Well 2 : 5µL of pSB1C3+1µL of 6X loading dye | ||
| + | * Well 3 : 5µL of RBS-BphR2 Part I+1µL of 6X loading dye | ||
| + | * Well 4 : 5µL of RBS-FNR Part I+1µL of 6X loading dye | ||
| + | * Well 5 : 5µL of FNR Part I+1µL of 6X loading dye | ||
| + | * Well 6 : 5µL of FNR Part II+1µL of 6X loading dye | ||
| + | * Well 7 : 5µL of BphR2 Part II+1µL of 6X loading dye | ||
| + | * Gel : 1% | ||
|} | |} | ||
| - | + | Expected sizes : | |
| - | + | * pSB1C3 : 2070 bp | |
| - | + | * RBS-BphR2 Part I : 197 bp | |
| - | + | * BphR2 Part II : 790 bp | |
| - | + | * RBS-FNR Part I : 615 bp | |
| - | + | * FNR Part I : 597 bp | |
| - | + | * FNR Part II : 200 bp | |
| - | + | ||
| - | + | {| | |
| - | | | + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | |
| - | + | We obtain fragment at the right size for RBS-BphR2 Part I, BphR2 Part II, RBS-FNR Part I, FNR Part I, FNR Part II but not for pSB1C3. We will do a digestion of pSB1C3 by DnpI to clean it. | |
| - | |- | + | |
| - | | | + | |
| - | + | ||
|} | |} | ||
| - | : | + | ===='''2 - Digestion of pSB1C3 by DnpI'''==== |
| + | |||
| + | XiaoJing | ||
| + | |||
| + | Used quantities : | ||
| + | * pSB1C3 : 17µL | ||
| + | * Buffer : 2µL | ||
| + | * DnpI : 1µL | ||
| + | |||
| + | We keep the digestion for 1h30 at 37°C. | ||
| + | |||
| + | ===='''3 - Electrophoresis of the digestion of pSB1C3 by DnpI'''==== | ||
| - | + | XiaoJing | |
| - | |||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" | [[]] | + | | style="width:350px;border:1px solid black;" | [[File:Psgel32708.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
| - | *Well 1 : 6µL DNA | + | * Well 1 : 6µL DNA ladder |
| - | *Well | + | * Well 3 : 20µL of pSB1C3 digested by DnpI + 4µL of 6X loading dye |
| - | * | + | * Gel : 1% |
| + | |} | ||
| + | Expected sizes : | ||
| + | * pSB1C3 : 2070 bp | ||
| - | + | {| | |
| + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| + | We obtain fragment at the right size for pSB1C3. We will purify it. | ||
|} | |} | ||
| - | |||
{| border="1" align="center" | {| border="1" align="center" | ||
| - | ||<big>Previous | + | |[[Team:Paris Saclay/Notebook/August/26|<big>Previous day</big>]] |
|[[Team:Paris_Saclay/Notebook|<big>Back to calendar</big>]] | |[[Team:Paris_Saclay/Notebook|<big>Back to calendar</big>]] | ||
| - | |[[Team:Paris Saclay/Notebook/ | + | |[[Team:Paris Saclay/Notebook/August/28|<big>Next day</big>]] |
|} | |} | ||
| - | |||
{{Team:Paris_Saclay/incl_fin}} | {{Team:Paris_Saclay/incl_fin}} | ||
Latest revision as of 01:20, 5 October 2013
Notebook : August 27
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000 and BBa_K1155004
1 - PCR Colony of ligation Pndh* or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in pSB1C3
XiaoJing
|
Transformation of 08/26/13 works. We will do a Colony PCR. |
We took a single colony and resuspend in 10µL H2O.For each Biobrick we did 6 PCR Colony.
Used quantities :
- DNA : 2µL
- Mix : (it was divided in 6 tubes for each promotor with 23µL of mix in each tube)
- Oligo 44 : 17.5µL
- Oligo 43 : 17.5µL
- Buffer Dream Taq : 87.5µL
- dNTP : 17.5µL
- Dream Taq : 7µL
- H2O : 591µL
PCR Program :
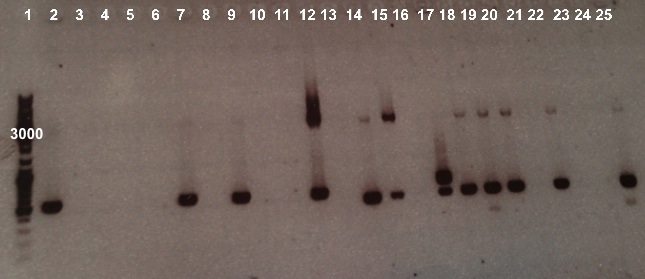
2 - Electrophoresis of the colony PCR products : Pndh* or NirB with RBS-LacZ-Term or RBS-Amil CP-Term in pSB1C3
XiaoJing
Expected size :
- NirB with RBS-LacZ-Term in pSB1C3 : 3474bp
- NirB with RBS-AmilCP-Term in pSB1C3 : 1029bp
- Pndh* with RBS-LacZ-Term in pSB1C3 : 3380bp
- Pndh* with RBS-AmilCP-Term in pSB1C3 : 935bp
|
We obtained fragments at the right size for NirB with RBS-Amil CP-Term in pSB1C3 in well 12, Pndh* with RBS-LacZ-Term in pSB1C3 in well 14, 15, 18 and 19 and Pndh* with RBS-Amil CP-Term in pSB1C3 in well 20, 22 and 25. Nevertheless, electrophoresis shows that these colonies weren't pure. We will purify them by streaking. |
3 - Streak colonies of NirB with RBS-Amil CP-Term in pSB1C3, Pfnr with RBS-LacZ-Term in pSB1C3 and Pfnr with RBS-Amil CP-Term in pSB1C3 to purify them
XiaoJing
4 - Culture of strain MG1655Z1 Δfnr::Km containing plasmid pcp20
XiaoJing
|
Purification from 08/26/13 works. |
We made culture on LB medium at 42°C for strain MG1655Z1 Δfnr::Km containing plasmid pcp20 to select the bacteria that eliminate the Km cassette. Therefore, we can use this strain to receive the plasmid IGEM pSB1K3. Like this we will select clone Δfnr::Km by streaking them on plate with ampicilin, kanamycin and chloramphenicol plates.
Principle :
Plasmid pcp20 is thermosensitive plasmid which produces a flipase that is able to eliminate antibiotic gene (in our case is Km) at 30°C. When we shift back the strain at 42°C, the plasmid can not replicate and the bacteria will loose their plasmid. As a result, we constructed successfully the mutant strain MG1655Z1::Δfnr without any antibiotic resistance.
A - Aerobic/Anaerobic regulation system / B - PCB sensor system
Objective : obtaining FNR, BphR2 proteins
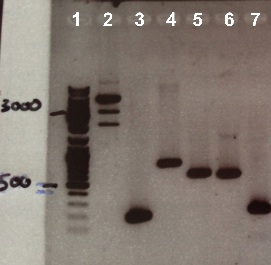
1 - Electrophoresis of RBS-BphR2 Part I, BphR2 Part II, FNR Part I , FNR Part II, RBS-FNR Part I and pSB1C3
XiaoJing
|
Gibson tranformation of the 08/27/13 didn't work. So we did an electrophoresis to check sizes and concentrations of Gibson parts. |
Expected sizes :
- pSB1C3 : 2070 bp
- RBS-BphR2 Part I : 197 bp
- BphR2 Part II : 790 bp
- RBS-FNR Part I : 615 bp
- FNR Part I : 597 bp
- FNR Part II : 200 bp
|
We obtain fragment at the right size for RBS-BphR2 Part I, BphR2 Part II, RBS-FNR Part I, FNR Part I, FNR Part II but not for pSB1C3. We will do a digestion of pSB1C3 by DnpI to clean it. |
2 - Digestion of pSB1C3 by DnpI
XiaoJing
Used quantities :
- pSB1C3 : 17µL
- Buffer : 2µL
- DnpI : 1µL
We keep the digestion for 1h30 at 37°C.
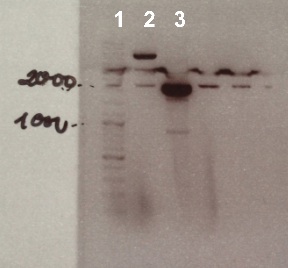
3 - Electrophoresis of the digestion of pSB1C3 by DnpI
XiaoJing
|
|
Expected sizes :
- pSB1C3 : 2070 bp
|
We obtain fragment at the right size for pSB1C3. We will purify it. |
| Previous day | Back to calendar | Next day |
 "
"