Team:TU-Eindhoven/ProteinSelection
From 2013.igem.org
Pascalaldo (Talk | contribs) |
Pascalaldo (Talk | contribs) |
||
| Line 5: | Line 5: | ||
{{:Team:TU-Eindhoven/Template:Lead}}To create bacteria with the ability to generate contrast on a CEST MRI scan, polypeptides that have that ability had to be found. Various solutions based on short Lysine, Arganine, Threonine or Serine rich sequences were proposed.{{:Team:TU-Eindhoven/Template:Ref | id=McMahonDIACEST | author=M.T. McMahon | title=New "Multicolor" Polypeptide Diamagnetic Chemical Exchange Saturation Transfer (DIACEST) Contrast Agents for MRI | journal=Magnetic Resonance in Medicine | edition=60 | pages=803-812 | year=2008 }} However, it is hard to predict how well these sequences will express in bacteria and whether they are sufficiently stable in vivo. To avoid these problems a new approach was taken. The suitability as a CEST based marker was estimated for proteins of which the structure is already clarified.{{:Team:TU-Eindhoven/Template:LeadEnd}} | {{:Team:TU-Eindhoven/Template:Lead}}To create bacteria with the ability to generate contrast on a CEST MRI scan, polypeptides that have that ability had to be found. Various solutions based on short Lysine, Arganine, Threonine or Serine rich sequences were proposed.{{:Team:TU-Eindhoven/Template:Ref | id=McMahonDIACEST | author=M.T. McMahon | title=New "Multicolor" Polypeptide Diamagnetic Chemical Exchange Saturation Transfer (DIACEST) Contrast Agents for MRI | journal=Magnetic Resonance in Medicine | edition=60 | pages=803-812 | year=2008 }} However, it is hard to predict how well these sequences will express in bacteria and whether they are sufficiently stable in vivo. To avoid these problems a new approach was taken. The suitability as a CEST based marker was estimated for proteins of which the structure is already clarified.{{:Team:TU-Eindhoven/Template:LeadEnd}} | ||
| - | + | ==Scanning for Candidates== | |
For good CEST contrast a protein should have a high Lysine or Arganine content.{{:Team:TU-Eindhoven/Template:RefAgain | id=McMahonDIACEST }}{{:Team:TU-Eindhoven/Template:Ref | id=GiladArtificialGene | author=A.A. Gilad | title=Artificial reporter gene providing MRI contrast based on proton exchange. | journal=Nature biotechnology | edition=25.2 | pages=217-219 | year=2007 }} To find these suitable proteins the [http://www.pdb.org RCSB Protein Data Bank] was queried for all entries containing proteins. This was done using the SEARCH Web Service of PDB.org, for the XML query used see [[Team:TU-Eindhoven/Code:PDBQuery#query.xml | query.xml]]. A Python program was written to analyse the obtained amino acid sequences and calculate the ratio of Lysine or Arginine to the total chain length (see [[Team:TU-Eindhoven/Code:PDBQuery#query.xml | PDB.py]] and [[Team:TU-Eindhoven/Code:PDBQuery#query.xml | queryPDB.py]]). The results are visualized in {{:Team:TU-Eindhoven/Template:Figure | id=lysineRatioPlot }} and {{:Team:TU-Eindhoven/Template:Figure | id=arginineRatioPlot }}. | For good CEST contrast a protein should have a high Lysine or Arganine content.{{:Team:TU-Eindhoven/Template:RefAgain | id=McMahonDIACEST }}{{:Team:TU-Eindhoven/Template:Ref | id=GiladArtificialGene | author=A.A. Gilad | title=Artificial reporter gene providing MRI contrast based on proton exchange. | journal=Nature biotechnology | edition=25.2 | pages=217-219 | year=2007 }} To find these suitable proteins the [http://www.pdb.org RCSB Protein Data Bank] was queried for all entries containing proteins. This was done using the SEARCH Web Service of PDB.org, for the XML query used see [[Team:TU-Eindhoven/Code:PDBQuery#query.xml | query.xml]]. A Python program was written to analyse the obtained amino acid sequences and calculate the ratio of Lysine or Arginine to the total chain length (see [[Team:TU-Eindhoven/Code:PDBQuery#query.xml | PDB.py]] and [[Team:TU-Eindhoven/Code:PDBQuery#query.xml | queryPDB.py]]). The results are visualized in {{:Team:TU-Eindhoven/Template:Figure | id=lysineRatioPlot }} and {{:Team:TU-Eindhoven/Template:Figure | id=arginineRatioPlot }}. | ||
{{:Team:TU-Eindhoven/Template:Float | position=left | size=6 }} | {{:Team:TU-Eindhoven/Template:Float | position=left | size=6 }} | ||
| Line 110: | Line 110: | ||
| 0.25 | | 0.25 | ||
|} | |} | ||
| - | + | ||
| + | ==Molecular Dynamics== | ||
To refine the selection the accessibility of the various exchangable hydrogen atoms of the Lysines and Arginines was taken into account. Hereto Molecular Dynamics simulations of the proteins in water were carried out. | To refine the selection the accessibility of the various exchangable hydrogen atoms of the Lysines and Arginines was taken into account. Hereto Molecular Dynamics simulations of the proteins in water were carried out. | ||
| - | + | ||
| + | ==References== | ||
{{:Team:TU-Eindhoven/Template:RefList}} | {{:Team:TU-Eindhoven/Template:RefList}} | ||
Revision as of 14:01, 2 July 2013



Contents |
CEST Based Marker Proteins
To create bacteria with the ability to generate contrast on a CEST MRI scan, polypeptides that have that ability had to be found. Various solutions based on short Lysine, Arganine, Threonine or Serine rich sequences were proposed.McMahonDIACESTM.T. McMahon, New "Multicolor" Polypeptide Diamagnetic Chemical Exchange Saturation Transfer (DIACEST) Contrast Agents for MRI. Magnetic Resonance in Medicine 60, 803-812 (2008) However, it is hard to predict how well these sequences will express in bacteria and whether they are sufficiently stable in vivo. To avoid these problems a new approach was taken. The suitability as a CEST based marker was estimated for proteins of which the structure is already clarified.
Scanning for Candidates
For good CEST contrast a protein should have a high Lysine or Arganine content.McMahonDIACESTGiladArtificialGeneA.A. Gilad, Artificial reporter gene providing MRI contrast based on proton exchange.. Nature biotechnology 25.2, 217-219 (2007) To find these suitable proteins the RCSB Protein Data Bank was queried for all entries containing proteins. This was done using the SEARCH Web Service of PDB.org, for the XML query used see query.xml. A Python program was written to analyse the obtained amino acid sequences and calculate the ratio of Lysine or Arginine to the total chain length (see PDB.py and queryPDB.py). The results are visualized in and .
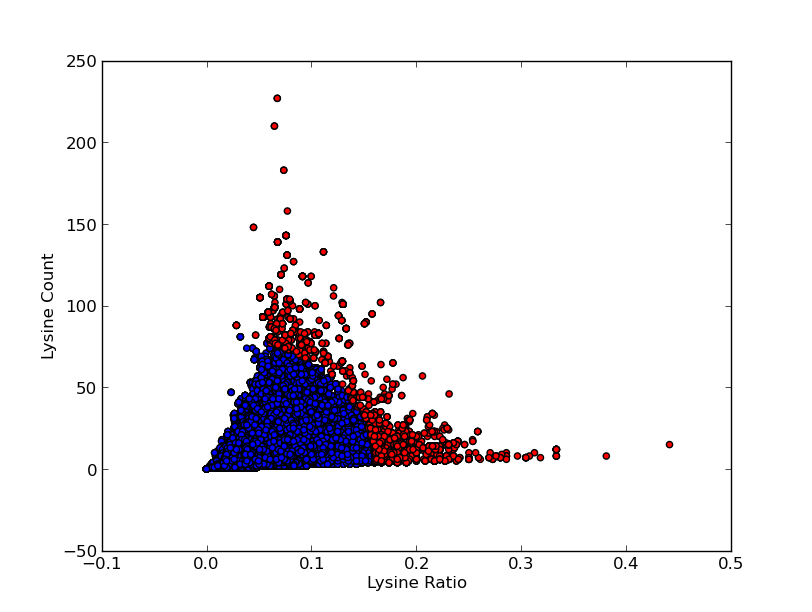
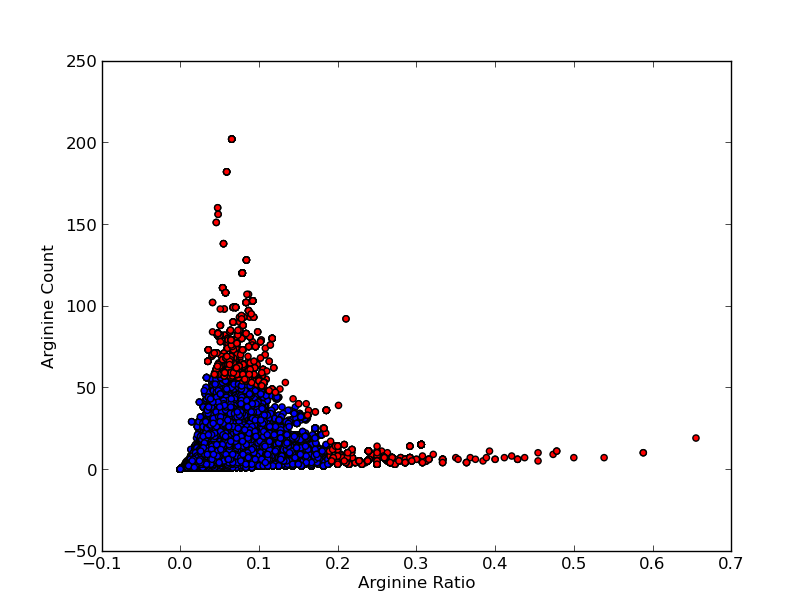
| PDB ID | Frequent Amino Acid | Amount of Frequent Amino Acids (#) | Chain Length (#) | Ratio (-) |
|---|---|---|---|---|
| 1ETF | Arginine | 11 | 23 | 0.48 |
| 1IWQ | Lysine | 7 | 19 | 0.37 |
| 1PJN | Lysine | 8 | 21 | 0.38 |
| 2IGR | Lysine | 15 | 34 | 0.44 |
| 2KLW | Lysine | 10 | 32 | 0.31 |
| 2PCO | Lysine | 8 | 26 | 0.30 |
| 1G70 | Arginine | 10 | 22 | 0.46 |
| 1BY0 | Lysine | 8 | 27 | 0.30 |
| 1NWD | Lysine | 8 | 28 | 0.29 |
| 1PEH | Lysine | 10 | 35 | 0.29 |
| 2L9A | Lysine | 8 | 24 | 0.33 |
| 2L96 | Lysine | 8 | 24 | 0.33 |
| 2L99 | Lysine | 8 | 24 | 0.33 |
| 1LYP | Lysine | 9 | 32 | 0.28 |
| 1LQ7 | Lysine | 17 | 67 | 0.25 |
Molecular Dynamics
To refine the selection the accessibility of the various exchangable hydrogen atoms of the Lysines and Arginines was taken into account. Hereto Molecular Dynamics simulations of the proteins in water were carried out.
References
 "
"