Team:TU-Munich/Results/GM-Moss
From 2013.igem.org
Creation of transgenic Physcomitrella patens plants
Generation of expression constructs
| Number | Construct name (abbreviation) | BioBrick | Figure | Size [bp] | Lane | GM-Moss |
|---|---|---|---|---|---|---|
| PF-1 | GFP (GFP-cyt) | <partinfo>BBa_K1159311</partinfo> | x | 
| ||
| PF-2 | Igk-GFP (GFP-sec) | <partinfo>BBa_E0040</partinfo> | x | 
| ||
| PF-3 | NanoLuciferase (nLuc-cyt) | <partinfo>BBa_K1159001</partinfo> | x | 
| ||
| PF-4 | Igk-NanoLuciferase (nLuc-sec) | <partinfo>BBa_K1159006</partinfo> | x | 
| ||
| PF-5 | SERK-NanoLuciferase (SERK-nLuc) | <partinfo>BBa_K1159010</partinfo> | x | 
| ||
| PF-6 | NanoLuciferase-Receptor (nLuc-rec) | <partinfo>BBa_K1159015</partinfo> | x | 
| ||
| PF-7 | Erythromycinesterase (EreB-cyt) | <partinfo>BBa_K1159000</partinfo> | x | 
| ||
| PF-8 | Erythromycinesterase-Receptor (EreB-rec) | <partinfo>BBa_K1159014</partinfo> | # | 
| ||
| PF-9 | Ig Kappa Erythromycinesterase (EreB-sec) | <partinfo>BBa_K1159005</partinfo> | # | 
| ||
| PF-10 | Laccase-Receptor (Lac-rec) | <partinfo>BBa_K1159016</partinfo> | # | 
| ||
| PF-11 | Ig Kappa Laccase (Lac-sec) | <partinfo>BBa_K1159007</partinfo> | # | 
| ||
| PF-12 | Catecholdioxygenase (XylE-cyt) | <partinfo>BBa_E0040</partinfo> | x | 
| ||
| PF-13 | DDT-dehydrochlorinase (GST-cyt) | <partinfo>BBa_E0040</partinfo> | # | 
| ||
| PF-14 | PP1-Receptor (PP1-rec) | <partinfo>BBa_K1159019</partinfo> | x | 
| ||
| PF-15 | FluA-Receptor (FluA-rec) | <partinfo>BBa_K1159017</partinfo> | x | 
| ||
| PF-16 | Stressinducible_Promoter-RFP (Stress) | <partinfo>BBa_E0040</partinfo> | # | 
| ||
| PF-17 | SpyCatcher-Receptor:Spytag-nLuc (Catcher:Tag-nLuc) | <partinfo>BBa_E0040</partinfo> | # | 
| ||
| PF-18 | SpyCatcher-Receptor:nLuc-Spytag (Catcher:nLuc-Tag | <partinfo>BBa_E0040</partinfo> | # | 
| ||
| PF-19 | SpyTag-Receptor:SpyCatcher-nLuc (Tag:Catcher-nLuc) | <partinfo>BBa_E0040</partinfo> | # | 
| ||
| PF-20 | SpyTag-Receptor:nLuc-SpyCatcher (Tag:nLuc-Catcher) | <partinfo>BBa_E0040</partinfo> | # | 
| ||
| PF-21 | Alcohol acetyltransferase I (Banana) | <partinfo>BBa_J45014</partinfo> | # | 
| ||
| PF-22 | Kill-switch with PIF3 (PIF-3) | <partinfo>BBa_E0040</partinfo> | x | no | ||
| PF-23 | Kill-switch with PIF6 (PIF-6) | <partinfo>BBa_E0040</partinfo> | x | no | ||
| PF-24 | Kill-switch with PIF6 (FRET) | <partinfo>BBa_E0040</partinfo> | # | 
| ||
| PF-25 | Kill-switch with PIF3 (FRET) | <partinfo>BBa_E0040</partinfo> | # | 
|
Preparation of linear DNA
Transformation of Physcomitrella patens
To transform Physcomitrella patens, the moss material has to be taken from the liquid culture and its cell walls have to be digested with driselase dissolved in mannitol to obtain protoplasts. The protoplasts are isolated by passing the digested material through sieves and the enzyme is washed off with mannitol and then resuspended in mannitol.
The number of protoplasts is determined with a hemocytometer and the material is suspended in the right amount of 3M medium to adjust the concentration. The linearized and purified DNA is mixed with PEG4000 and the protoplast solution and incubated while regularly mixing. After incubation, the mixture is diluted with 3M medium, centrifuged and resuspended in regeneration medium.
The protoplasts are put into 6-well plates, left in the dark over night and then left for 10 days for the regeneration of the cell walls at standard conditions. After moving the protoplasts onto solid medium covered with a layer of cellophane for three days, they are transferred to solid selection medium plates for two weeks. To ensure stabile integration, repeat the two weeks of selection after a two week release phase.
Trips to Freiburg
We had the great chance to perform our Physcomitrella patens transformations at Prof. Dr.Reski´s lab in Freiburg, with Dr. Wiedemann as our expert instructor. Our first trip started in a great hurry, because we worked on our DNA preparations until the very last minute. Ingmar even pulled an all-nighter to get the DNA ready. We would have missed our intercity bus if it wasn’t for Rosario who drove us to the bus station, all squished together in the “pizza mobile” with the trunk full with our medium bottles and lab equipment. After five hours on the bus, we fell into bed to get some rest, because we had a very long day ahead of us.
We arrived at the Reski lab early in the morning to meet with Dr. Gertrud Wiedemann, who instructed us throughout the day and gave us many tips how to proceed with the moss. Because of the incubation times and because it was our first try, it took us ten hours without a break until we had two boxes stacked with 6 well plates. We quickly went to get some beers and chips and met with the Freiburg iGEM team for a really nice barbecue. When we finally left, public transport wasn’t running anymore, so we didn’t miss the chance to take a midnight sightseeing tour through Freiburg, where Volker showed us around.
Ten days later, Johanna and Andi visited the Freiburg lab to transfer the then regenerated protoplasts onto agar plates and soon after, we came back for our second and final round of transformation. At our first try, we didn’t get enough moss protoplasts, so we worked through two batches and therefore had to prepare another batch of driselase. Our handling had improved, yet it still took twelve hours and again, there was no time left for a break. After we said good bye, we celebrated with a couple of beers and some yummy flammkuchen at the UC uni café of Freiburg. We had learned so much and got much closer to our goal. A big successful step for our team!
Regeneration and Selection of transgenic plants
Characterization of transgenic plants
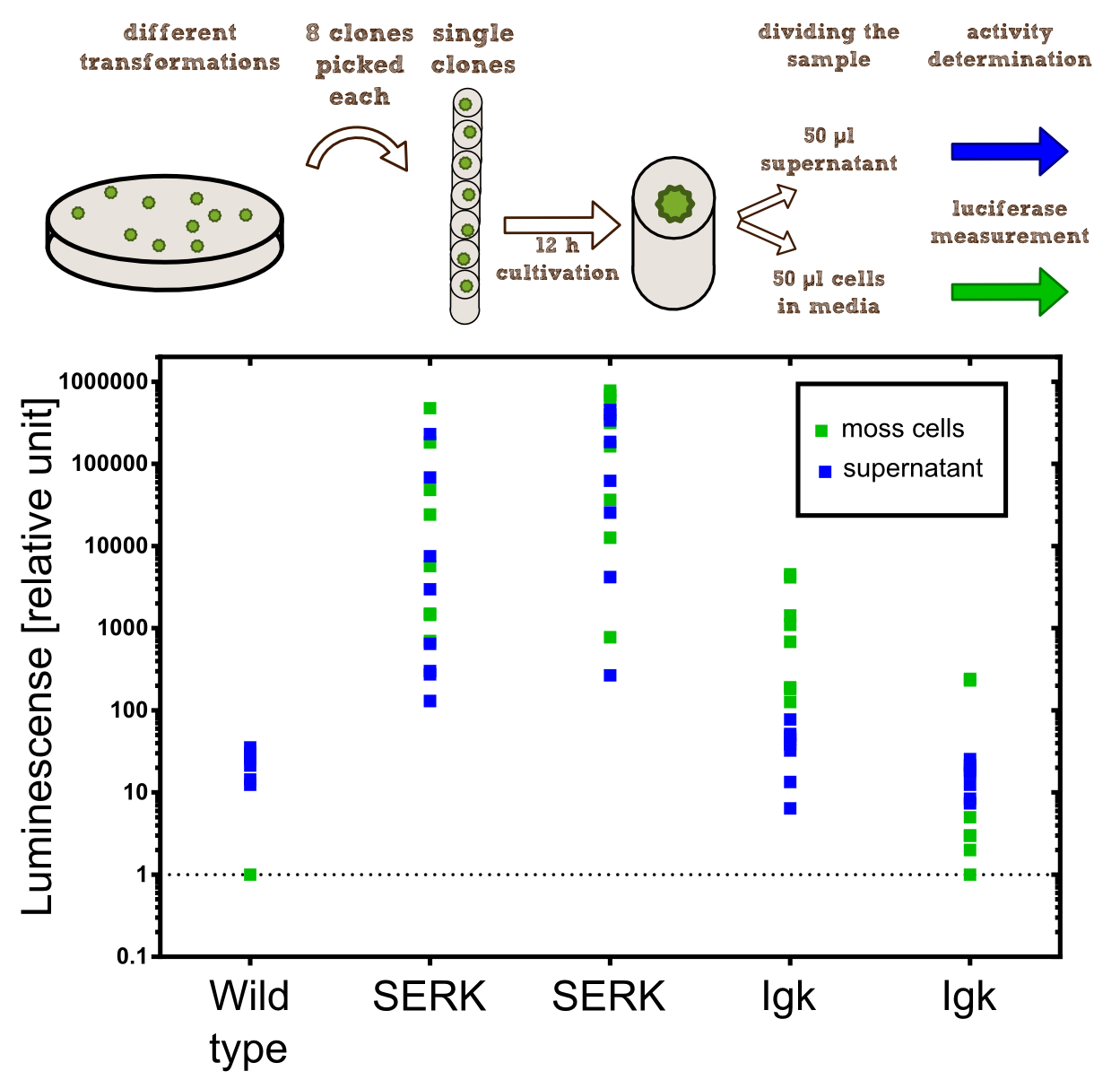
Investigation of the localization within the moss cell
Idea We want to show that our membrane construct, extracellular containing Nanoluciferase fused to a Streptag for purification and a TEV split site to cut of the Nanoluciferase-Streptag-Construct, is expressed and afterwards stocked in the membrane of our moss.
To prove this process, we want to cut of our Nanoluciferase-Streptag-Construct by using the TEV protease. As we hypothetically know, our membrane constructs are localized in the membrane, that means protected by the Cell wall of our moss.In conclusion to that, the TEV protease needs to pass the cell wall to cut of the Nanoluciferase. This process is possible in liquid medium, because the TEV protease just has a molecular weight between 25 kDa and 27 kDa. After the TEV protease has entered the Periplasma of the moss cell, it cuts off the Nanoluciferase-Streptag-Construct at the TEV cleavage site. Again, this construct just has in sum a molecular weight of 20 kDa (Nanoluciferase 19 kDa, Streptag 1 kDa), so it can pass the cell wall to the exterior. This process takes place because of diffusion.
After this experimental part, it is necessary to prove the theoretical deliberation. So we performed several experiments like SDS-Page plus Western Blot using different antibodies, Luciferase Assays and tried to bind our construct by utilizing different beads.
Experimental Setup for localization process of membrane bound Nanoluciferase
First Step
1,5 ml TEV Buffer 100µl TEV Moss particles as much as possible - Nanoluc membrane bound Incubation over night
Second Step
Centrifugation 5 min 13000 rpm Supernatant in new Eppi Adding 50 µl of 1 mg/ml Biotin Beads Incubation over night
NK is Biotin Beads + PBS
Third step
Centrifugation of beads Supernatant of PK Plate Reader - 3 Reads No bond nanoluc Just in supernatant
Fourth step
concentration of Supernatant to 150 µl
Fifth step
3x 15 µl Supernatant + 7 µl red. SDS SDS-Page is it abundant?
Sixth step
Western Blot
Investigation of different Signal peptides for secretion of effector proteins
Characterization of intracellular localization of recombinant proteins
References:
http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984
- http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984 Edens, L., Bom, I., Ledeboer, A. M., Maat, J., Toonen, M. Y., Visser, C., and Verrips, C. T. (1984). Synthesis and processing of the plant protein thaumatin in yeast. Cell, 37(2):629–33.
- http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf Hohe, A., T. Egener, J. Lucht, H. Holtorf, C. Reinhard, G. Schween, R. Reski (2004): An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene knockouts in a moss, Physcomitrella patens. Current Genet. 44, 339-347.
 "
"











































AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351