Team:Uppsala/zeaxantin
From 2013.igem.org
| (29 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Uppsala/standard-style}} | {{Team:Uppsala/standard-style}} | ||
| - | {{Team:Uppsala/ | + | {{Team:Uppsala/zeaxanthin-style}} |
<html> | <html> | ||
| Line 27: | Line 27: | ||
<!-- Contains sub-logo picture --> | <!-- Contains sub-logo picture --> | ||
| - | <div id="igem_logo"><a href="https://2013.igem.org/ | + | <div id="igem_logo"><a href="https://2013.igem.org/Team:Uppsala"><img class="igem_logo" src="https://static.igem.org/mediawiki/2013/e/e0/Uppsala2013_IGEMlogo.png"></a></div> |
| - | <div id="igem"><a href="https://2013.igem.org/Main_Page"><img class="igem" src="https://static.igem.org/mediawiki/2013/ | + | <div id="igem"><a href="https://2013.igem.org/Main_Page"><img class="igem" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_Uppsala_IGEM_log_blue.png"></a></div> |
</div> | </div> | ||
| Line 44: | Line 44: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/project" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/a/aa/Uppsala2013_Project.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/project" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/a/aa/Uppsala2013_Project.png"></a> | ||
<ul> | <ul> | ||
| - | + | ||
<li><a href="https://2013.igem.org/Team:Uppsala/chassi">Chassi</a> | <li><a href="https://2013.igem.org/Team:Uppsala/chassi">Chassi</a> | ||
<ul> | <ul> | ||
| Line 50: | Line 50: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/promoters">Promoters</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/promoters">Promoters</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/reporter-genes">Reporter genes</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/reporter-genes">Reporter genes</a></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/ | + | <li><a href="https://2013.igem.org/Team:Uppsala/toxin-antitoxin-system">Toxin-antitoxin system</a></li> |
<li><a href="https://2013.igem.org/Team:Uppsala/vectors">Vectors</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/vectors">Vectors</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/signal-peptide">Signal peptide</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/signal-peptide">Signal peptide</a></li> | ||
| Line 64: | Line 64: | ||
href="https://2013.igem.org/Team:Uppsala/saffron">Saffron</a></li> | href="https://2013.igem.org/Team:Uppsala/saffron">Saffron</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/astraxantin">Astaxanthin</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/astraxantin">Astaxanthin</a></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/ | + | <li><a href="https://2013.igem.org/Team:Uppsala/zeaxanthin">Zeaxanthin</a></li> |
</ul></li> | </ul></li> | ||
</li> | </li> | ||
| Line 71: | Line 71: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/chromoproteins">Chromoproteins</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/chromoproteins">Chromoproteins</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/safety-experiment">Safety experiment</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/safety-experiment">Safety experiment</a></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/result"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/result">Results</a></li> |
| Line 88: | Line 88: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | ||
| - | + | ||
</ul></li> | </ul></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/hp" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_HP.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/hp" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_HP.png"></a> | ||
| Line 95: | Line 95: | ||
<li><a href="https://2013.igem.org/Team:Uppsala/synbioday">SynBioDay</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/synbioday">SynBioDay</a></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/safety">Biosafety and ethics</a></li> | <li><a href="https://2013.igem.org/Team:Uppsala/safety">Biosafety and ethics</a></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/society"> | + | <li><a href="https://2013.igem.org/Team:Uppsala/society">Public opinion </a></li> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/ | + | <li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> |
| - | + | ||
</ul></li> | </ul></li> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a> |
| + | <ul> | ||
| + | <li><a href="https://2013.igem.org/Team:Uppsala/collaboration">Collaboration</a></li> | ||
| + | </ul></li> | ||
<li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | <li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | ||
<ul> | <ul> | ||
| - | <li><a href="https://2013.igem.org/Team:Uppsala/ | + | <li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> |
</ul></li> | </ul></li> | ||
</ul> | </ul> | ||
| Line 111: | Line 114: | ||
<div id="main_content"> <!-- Put content here --> | <div id="main_content"> <!-- Put content here --> | ||
| - | <h1> | + | <h1 class="main-title-left">Zeaxanthin</h1> |
| - | + | <img class="metabolic-pic" src="https://static.igem.org/mediawiki/2013/e/e5/Uppsala2013_Zeaxantin2symbol.png"> | |
| - | + | <div id="clear"></div> | |
| - | <p> | + | |
| - | + | <p>Zeaxanthin is a yellow carotenoid pigment, derived from the precursor ß-carotene through hydroxylation by the enzyme ”Beta-carotene hydroxylase”. Zeaxanthin acts as an antioxidant and can be found in for example peppers, yolk and maize.<sup><a href="#l1">(1)</a></sup> According to studies zeaxanthin has positive effects on both undamaged and impaired vision and it may prevent age-related macular degeneration (AMD), an eye condition that could lead to blindness. Furthermore studies have also indicated that zeaxanthin may have skin protective activities<sup><a href="#l2"> (1,4).</a></sup> | |
| - | <h1> | + | <br><br> |
| - | <p> | + | |
| + | Except for having beneficial properties the carotenoid the carotenoid zeaxanthin is one of the primary precursors for the production of the <a href="https://2013.igem.org/Team:Uppsala/saffron">saffron metabolites</a> picrocrocin, crocin and safranal. The production of saffron was inspired by the work of Washington team <a href="https://2012.igem.org/Team:WashU">WashU iGEM 2012.</a> Since the gene (CrtZ) that was used for the production of zeaxanthine was a eukaryotic version from Arabidopsis Thaliana, we decided to focus on finding a suitable gene to optimize the production of the above-mentioned metabolites. When searching for an appropriate gene we came in contact with Slovenia iGEM 2010 (lank) that had produced Zeaxanthin in E.coli. The operon that was obtained contained genes that were originally from the bacteria <a href="http://parts.igem.org/Part:BBa_K323122">Pantoea Anantis.</a></p> | ||
| + | |||
| + | <a id="b1"></a><h1>Methods:</h1> | ||
| + | <p>For the characterization of zeaxanthin in E.coli DH5alpha we put together a standardized method for zeaxanthin extraction and measuring. Since zeaxanthin in its pure form is extremely expensive (~ 480 €/mg) we bought maize at our local supermarket and carried out liquid-liquid extractions using methanol and performed spectrophotometry measurements. We compared literature absorbance values of zeaxanthin to the peaks we obtained from our own measurements and continued our experiments until we believed to have an optimized extraction method.</p> | ||
| + | |||
| + | <img class="zeaxanthin1" src="https://static.igem.org/mediawiki/2013/9/9d/Uppsala2013_Zeaxanthin1.png"> | ||
| + | |||
| + | <i><b>Figure 1.</b> Maize bought at our local supermarket that was grinded and later used to make a standardized zeaxanthin extraction method </i> | ||
| + | <br> | ||
| + | <i><b>Figure 2.</b> Extraction experiments carried out on grinded maize using methanol.</i> | ||
| + | <br><br> | ||
| + | |||
| + | <p>The operon acquired from Slovenia iGEM 2010 consists of the genes CrtE, CrtB, CrtI, CrtY and CrtZ connected through zinc fingers and linkers and assembled with the inducible promoter pBAD/AraC. In our project the zinc fingers were not put into a lot of consideration. | ||
| + | <br><br> | ||
| + | |||
| + | Since we would eventually like to produce zeaxanthin in Lactobacillus in yoghurt it would be preferable to have a constitutive promoter, such as the CP-promoters. Parallel to the characterization of zeaxanthin in E.coli we therefore attempted to remove the pBAD/AraC promoter from the zeaxanthin operon.</p> | ||
| + | |||
| + | <img class="methods-plasmid" src="https://static.igem.org/mediawiki/2013/6/68/Uppsala2013_Zeaxanthin_photoshop_plasmid_andmolecules.png"> | ||
| + | |||
| + | <h1>Results:</h1> | ||
| + | <h3>Spectrophotometry</h3> | ||
| + | <p>Production of zeaxanthin in e-coli DH5alpha was detected trough spectrophotometry measurements performed after liquid-liquid extractions using methanol. The resulting absorbance curve was compared to the standardized curve. In both graphs you can find the two peaks corresponding to zeaxanthin.</p> | ||
| + | |||
| + | <img class="zeaxanthin3" src="https://static.igem.org/mediawiki/2013/3/30/Uppsala2013_zeaxanthin2.png"> | ||
| + | |||
| + | |||
| + | <i><b>Figure 3.</b> E.coli culture believed to have zeaxanthin production</i> | ||
| + | <br> | ||
| + | <i><b>Figure 4.</b> A liquid-liquid extraction was carried out on the E.coli culture containing the plasmid above. Methanol was used as an organic solvent.</i> | ||
| + | |||
| + | <div id="pic-box3"> | ||
| + | <img class="zeaxanthin5" src="https://static.igem.org/mediawiki/2013/0/04/Uppsala2013_Zeaxanthin5.jpg"> | ||
| + | <img class="zeaxanthin6" src="https://static.igem.org/mediawiki/2013/0/0e/Uppsala2013_Zeaxanthin6.jpg"> | ||
| + | </div> | ||
| + | |||
| + | <div id="clear"></div> | ||
| + | |||
| + | <i><b>Figure 5.</b> Spectrophotometry of zeaxanthin extraction from maize resulted in the graph above. The two peaks are characteristic for zeaxanthin.</i> | ||
| + | <br> | ||
| + | <i><b>Figure 6.</b> Spectrophotometry of an E.coli culture containing the plasmid with the zeaxanthin operon. The characteristic peaks of zeaxanthin are present, indicating that zeaxanthin was present in the sample.</i> | ||
| + | |||
| + | |||
| + | <br><br> | ||
| + | <p>Spectrophotometry measurements were also done on unmodified E.coli, as a negative control. These measurements did not result in the peaks that are characteristic for zeaxanthin. </p> | ||
| + | |||
| + | <img class="zeaxanthin7" src="https://static.igem.org/mediawiki/2013/d/d9/Uppsala2013_Zeaxanthin7.jpg"> | ||
| + | |||
| + | <i class="fig-text7"><b>Figure 7.</b> Spectrophotometry of unmodified E.coli did not result<br> in the peaks characteristic for zeaxanthin. </i> | ||
| + | |||
| + | <div id="clear"></div> | ||
| - | |||
| - | |||
</div> <!-- main_content ends --> | </div> <!-- main_content ends --> | ||
Latest revision as of 20:26, 3 October 2013
Zeaxanthin

Zeaxanthin is a yellow carotenoid pigment, derived from the precursor ß-carotene through hydroxylation by the enzyme ”Beta-carotene hydroxylase”. Zeaxanthin acts as an antioxidant and can be found in for example peppers, yolk and maize.(1) According to studies zeaxanthin has positive effects on both undamaged and impaired vision and it may prevent age-related macular degeneration (AMD), an eye condition that could lead to blindness. Furthermore studies have also indicated that zeaxanthin may have skin protective activities (1,4).
Except for having beneficial properties the carotenoid the carotenoid zeaxanthin is one of the primary precursors for the production of the saffron metabolites picrocrocin, crocin and safranal. The production of saffron was inspired by the work of Washington team WashU iGEM 2012. Since the gene (CrtZ) that was used for the production of zeaxanthine was a eukaryotic version from Arabidopsis Thaliana, we decided to focus on finding a suitable gene to optimize the production of the above-mentioned metabolites. When searching for an appropriate gene we came in contact with Slovenia iGEM 2010 (lank) that had produced Zeaxanthin in E.coli. The operon that was obtained contained genes that were originally from the bacteria Pantoea Anantis.
Methods:
For the characterization of zeaxanthin in E.coli DH5alpha we put together a standardized method for zeaxanthin extraction and measuring. Since zeaxanthin in its pure form is extremely expensive (~ 480 €/mg) we bought maize at our local supermarket and carried out liquid-liquid extractions using methanol and performed spectrophotometry measurements. We compared literature absorbance values of zeaxanthin to the peaks we obtained from our own measurements and continued our experiments until we believed to have an optimized extraction method.
 Figure 1. Maize bought at our local supermarket that was grinded and later used to make a standardized zeaxanthin extraction method
Figure 1. Maize bought at our local supermarket that was grinded and later used to make a standardized zeaxanthin extraction method
Figure 2. Extraction experiments carried out on grinded maize using methanol.
The operon acquired from Slovenia iGEM 2010 consists of the genes CrtE, CrtB, CrtI, CrtY and CrtZ connected through zinc fingers and linkers and assembled with the inducible promoter pBAD/AraC. In our project the zinc fingers were not put into a lot of consideration.
Since we would eventually like to produce zeaxanthin in Lactobacillus in yoghurt it would be preferable to have a constitutive promoter, such as the CP-promoters. Parallel to the characterization of zeaxanthin in E.coli we therefore attempted to remove the pBAD/AraC promoter from the zeaxanthin operon.

Results:
Spectrophotometry
Production of zeaxanthin in e-coli DH5alpha was detected trough spectrophotometry measurements performed after liquid-liquid extractions using methanol. The resulting absorbance curve was compared to the standardized curve. In both graphs you can find the two peaks corresponding to zeaxanthin.
 Figure 3. E.coli culture believed to have zeaxanthin production
Figure 3. E.coli culture believed to have zeaxanthin production
Figure 4. A liquid-liquid extraction was carried out on the E.coli culture containing the plasmid above. Methanol was used as an organic solvent.
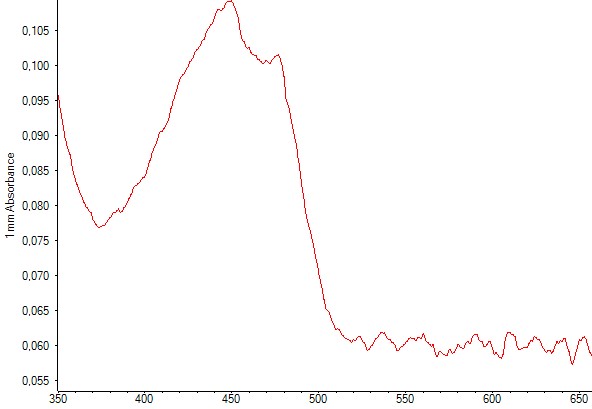

Figure 6. Spectrophotometry of an E.coli culture containing the plasmid with the zeaxanthin operon. The characteristic peaks of zeaxanthin are present, indicating that zeaxanthin was present in the sample.
Spectrophotometry measurements were also done on unmodified E.coli, as a negative control. These measurements did not result in the peaks that are characteristic for zeaxanthin.
 Figure 7. Spectrophotometry of unmodified E.coli did not result
Figure 7. Spectrophotometry of unmodified E.coli did not resultin the peaks characteristic for zeaxanthin.

 "
"









