Team:Duke/Project/Basics
From 2013.igem.org
Contents |
Background Information
Synthetic Gene Circuits and Bistable Switches
A key feature of synthetic biology is the “bottom-up” design of biological systems using existing biological parts or through the development of novel parts. Of particular interest in the field of synthetic biology is the understanding and development of genetic networks and engineering them to elicit a specific response. Given the dynamic nature of gene-protein interactions that can be explained by chemical kinetics and mathematical modeling, synthetic biologists can utilize these tools to design novel gene circuits guided by quantitative predictions. In 2000, three separate articles detailed the rational design of a genetic toggle switch (Gardner, 2000), an oscillatory circuit called the repressilator (Elowitz & Leibler, 2000), and an autoregulatory feedback circuit (Becskei & Serrano, 2000) in Escherichia coli. Since then, the regulatory motifs created through synthetic gene circuits have been expanded and combined to generate new responses such as logical computing or to improve the existing modules, such as robust oscillators.
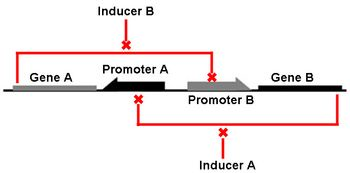
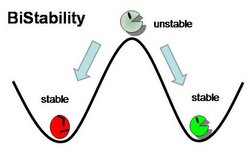
We focus our attention on the genetic toggle switch developed by Gardner, Cantor, and Collins in 2000, as this has guided the design of our project. The genetic toggle switch consists of two repressor proteins λ cI-ts, a temperature sensitive protein, and lacI, the lac repressor, linked in mutual repression in E. coli. The toggling comes from the presence of certain stimuli that turns on or off the production of one of the repressors. If the temperature is high (42 degrees Celsius), then λ cI-ts would be turned on and repress the production of lacI. This could be switched by changing the stimulus to IPTG, a chemical inducer that mimics allolactose to turn on the lacI gene and shut off the λ cI-ts gene. The ability to solely exist in one of two stable genetic states is a hallmark of bistability. The mutual repression of gene products generates a positive feedback loop, which exhibits bistability, with two stable steady states separated by an unstable steady state. By stable, we refer to the ability to make small perturbations to the system and have it return to the same state, and by unstable we refer to any small perturbation pushing the system away from the state into a more stable state. This can be demonstrated mathematically from the ordinary differential equations describing the chemical kinetics of gene expression or graphically by observing where the nullclines (where the derivative of gene expression is equal to zero) intersect, hinting at two stable intersections (steady states) separated by one unstable intersection.
Bistability and toggle switches serve valuable functions in biological systems in order to determine which genetic state a system should exist in response to external or internal stimuli. During stem cell differentiation, bistability is important as a cell decides which cell type to differentiate into, or to stay as a stem cell. The fate a cell makes is the result of toggling master control genes that lead to differentiated state or allowing the expression of pluripotentcy genes while silencing other cell type-specific genes. The same goes for cell division, where a cell can toggle between a quiescent state and a dividing state by controlling the expression of cell cycle genes such as cyclin-dependent kinases (Tanouchi, 2009). The ability to design bistable toggle switches also has applications to controlling gene expression in biotechnology production and gene therapies.
TAL Effector
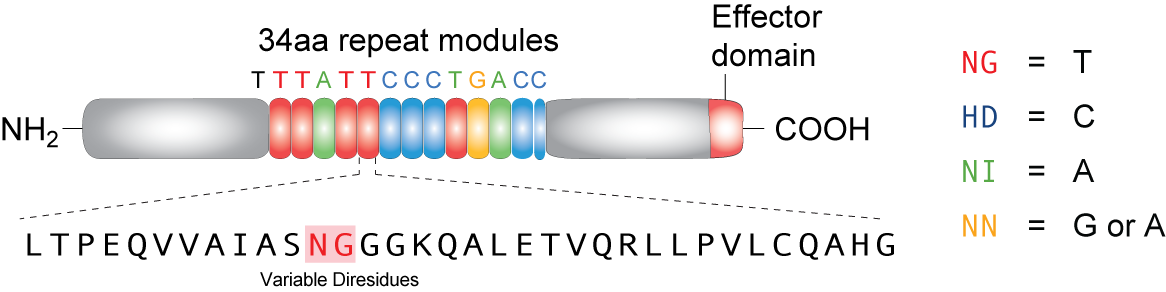
TALEs (Transcription Activator-Like effectors or TAL effectors) are DNA binding proteins derived from a plant pathogen Xanthomonas sp. which uses these proteins to regulate the host plant’s genome and aid in bacterial infection. Biochemical analysis of the TALEs revealed a highly repetitive primary structure with two residues at positions 12 and 13 out of a 33-34 amino acid protein that varied based on the target nucleotide. These two amino acids, referred to as repeat variable di-residues (RVDs) can be selected to target any DNA sequence and linked together to bind sequences up to 30 nucleotides long (Cermak et al., 2011). The four RVDs that we utilize are NG targeting T, HD targeting C, NI targeting A, and NN targeting G, although other RVDs exist that target various nucleotides with varying affinity (Mak et al., 2012). Using Golden Gate cloning, we are able to rapidly synthesize TALEs for uses in regulating gene expression by fusing activator or repressor domains to the TALEs. In addition, TALEs have been fused to nuclease domains (TALENs) for applications in genetic engineering and genome editing in plant and mammal cells by editing specific mutations, knocking out genes, or adding genes. We utilize a TALE fused to the fluorescent protein mCherry to act as a transcriptional roadblock rather than a repressor protein with the hopes of achieving the same result.
CRISPR
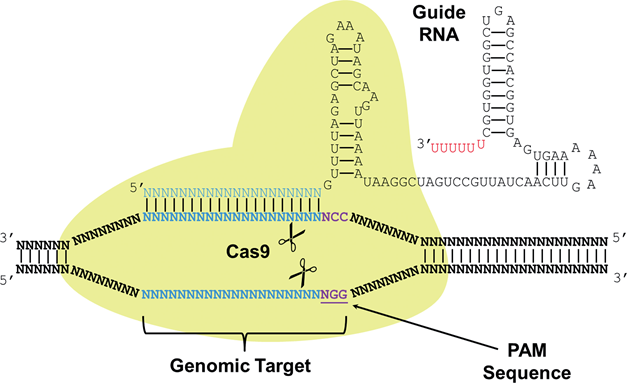
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) are a part of a system joined with Cas (CRISPR associated) proteins that function as a immune system found in around 90% of archaea and 40% of bacteria species. CRISPR-Cas systems recognize foreign DNA sequences and silence them through interactions with Cas proteins that have nuclease or helicase activity (DiCarlo et al., 2013). A small RNA called sgRNA (small-guide RNA) recognizes a specific DNA sequence and contains a region of significant secondary structure called the Cas9 handle that Cas proteins bind to either cut the DNA sequence or silence it. The use of the CRISPR-Cas9 system as a tool for genetic engineering is recent and rapidly growing because of the ability to multiplex the genome editing process by designing multiple sgRNAs targeting various regions in the genome to be edited simultaneously in conjunction with the nuclease activity of Cas9. Researchers have also discovered two nonsynonymous amino acid substitutions that deactivate the nuclease activity of Cas9 while preserving the binding activity of Cas9 to sgRNAs (Qi et al., 2013). This catalytically dead Cas9 is referred to as dCas9 and can be utilized in a similar fashion to TALEs where sgRNAs target promoters and the dCas9 protein acts as a roadblock to transcriptional activation. Recently, activating and repressing domains were fused to dCas9 to develop a gene regulation CRISPR/dCas9 system (Perez-Pinera et al., 2013). We utilize the dCas9 protein in order to cause a transcriptional roadblock for our toggle switch, very much like a repressor protein.
References
- Gardner T, Cantor C, Collins J: Construction of a genetic toggle switch in Escherichia coli. Nature 2000. 403:339-342.
- Elowitz, M., Liebler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000. 403, 335-228.
- Becskei, A., Serrano, L. Engineering stability in gene networks by autoregulation. Nature 2000. 405, 590-593.
- Tanouchi, Y., Pai, A., & You, L. Decoding biological principles using gene circuits. Mol Biosyst 2009. 5, 695-703.
- Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 2011. 39, e82.
- Mak A, Bradley P, Cernadas R, Bodfanove A, Stoddard B: The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 2012. 335, 716-719.
- DiCarlo J, Norville J, Mali P, Rios X, Aach J, Church M: Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Research 2013. 41(7), 4336-4343.
- Qi, LS., et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013. 152, 1173-1183.
- Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods 2013. 10, 973-976.
 "
"