The lac repressor/operator system uses E.coli to regulate the production of enzymes and because of this E. coli also regulate its metabolic stress. Enzymes only are produced when they are required. LacI is a repressor which is able to inhibit the lac operon in the absence of lactose by binding to the DNA at the lac operator site called lacO. Because of this repressor DNA polymerase is inhibited so it can’t read the sequences behind the operator lacZ (β-galactosidase), lacY (lactose permease) and lacA (thiogalactosidase transacetylase) which are responsible for transporting and metabolism of lactose in E. coli can’t be transcribed. The structure of the lac operon is shown in the image below.

Figure 1:Structure of the lactose operon and regulatory units.
When L-rhamnose is in the milieu where E. coli is located it can be taken up by the RhaT transport system which converts it to L-rhamnulose by the isomerase RhaA. It continues by phosphorylating by the kinase RhaB in rhamnulose-1-phosphate. This is hydrolyzed by the aldolase RhaD into dihydroxyacetone phosphate and lactate aldehyde. Dihydroxyacetone is metabolized in glycolysis, and lactate aldehyde aerobe to lactate. If there are anaerobe conditions lactate aldehyde is reduced to L-1,2,-propandiol. The gene RhaBAD functions as an operon and is transcribed by RhaPBAD. Two activators, RhaR and RhaS, have to be expressed to regulate the system. This expression of these activators is in opposite direction than the expression of rhaBAD. When L-rhamnose is available RhaR binds to RhaPRS and activates the production of RhaR and RhaS. RhaS binds with L-rhamnose as an effector to RhaPBAD and RhaPT promoter and activates the transcription of the structural genes.

L-rhamnose inducible expression system.
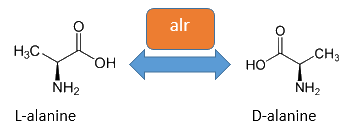
Alanine racemase is an isomerase which uses a covalently-bound pyridoxal 5’-phosphate (PLP) cofactor that catalyzes the racemization from L-alanine to D-alanine. D-alanine appears in peptidoglycan which is an important component of the bacterial cell wall.
The Barnase (EC 3.1.27) is an 12 kDa extracellular microbial ribonuclease, which is naturally found in the gram-positive soilbacteria Bacillus amyloliquefaciens and consist a single chain of 110 amino acids. The Barnase (RNase Ba) catalyses the cleavage of single stranded RNA, where the hydrolysis of the dinucleotides has the highest affinity to the structure GpN. In the first step of the RNA-degradation a cyclic intermediate is formed by transesterification and afterwards this intermediate is hydrolysed yielding in a 3'-nucleotide (see graphic below).
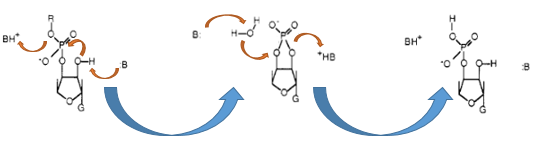
Figure x: Chemical reaction of ht e RNA-cleavage by the RNase Ba. First the transesterifiaction by the Glu-73 residue is performed and then this cyclic intermediat is hydrolized by the His-102 of the Barnase
References:
Agnes Ullmann (2001): Escherichia coli Lactose Operon. In: Encyclopedia of Life Sciences
Stumpp et al.: Ein neues, L-Rhamnose-induzierbares Expressionssystem für Escherichia coli, In: Biospektrum 6. Jahrgang S. 33
Carsten Voss, Dennis Lindau, and Erwin Flaschel, Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use, Biotechnology Progress, 22, 2006 p. 737-44.
Danuta E. Mossakowska, Kerstin Nyberg, and Alan R. Fersht, Kinetic Characterization of the Recombinant Ribonuclease from Bacillus amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis, Biochemistry, 28, 1989, p. 3843 – 3850.
C. J. Paddon, N. Vasantha, and R. W. Hartley, Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase, Journal of Bacteriology, 171, 1989, p. 1185 – 1187.
 "
"