Team:Bielefeld-Germany/Collaborations
From 2013.igem.org
Collaborations
Abstract
Beside the research in the own project's field it is important to collaborate with other iGEM teams in order to share knowledge and face challenges together. In this way, ideally both teams can profit from the work and experiences from one another and push their individual projects even further. In concrete terms, we sought to support various teams in their advances, for example by designing measurement systems for them or by submitting to them experimental data they required. In the following we list our efforts towards this.
iGEM-Team York_UK
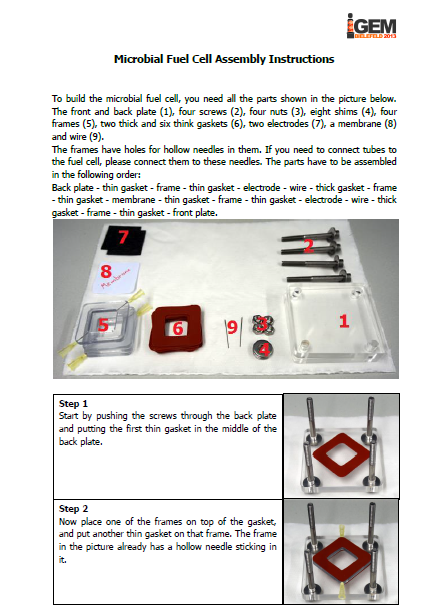
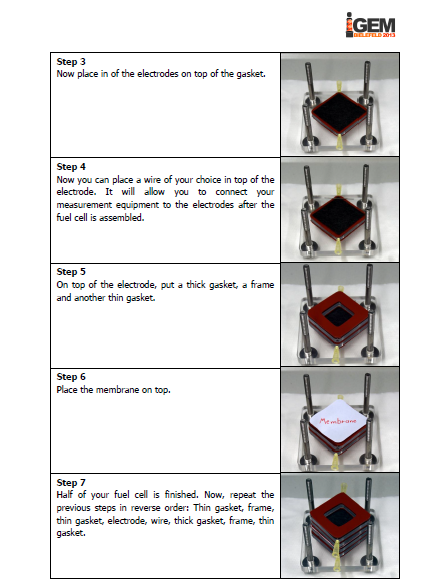
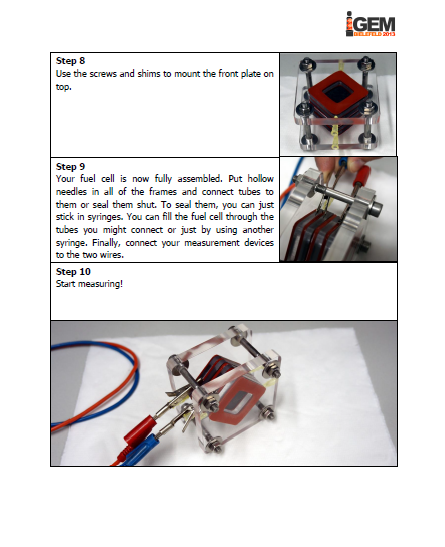
The iGEM-team York did, similar to our project, research into microbial fuel cells and so we got in touch and talked about our proceedings. We came to know that York had problems cloning the mtrCAB cluster and invested a lot of time to solve these problems. Time was certainly running at that point and we decided to help York. We already designed our MFC and tested different constructions in order to determine which one would be the most suitable. With all the experience we collected with MFC-construction, we were able to construct a MFC specially adapted to the requirements of team York. To ease up the usage of this cell, we wrote a detailed instruction on how to operate the MFC. Additionally we put nafion membranes, gaskets, carbon electrodes and injectors into a package and sent it to York. We hope that this support helped York to generate good results and finish their project.
Figure 1: We sent the instruction manual for our customized microbial fuel cell to team York along membranes, gaskets, carbon electrodes and injectors. |
iGEM-Team NRP-UEA-Norwich
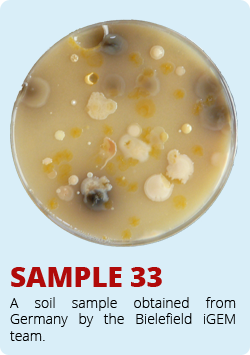
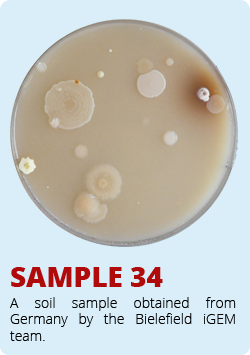
The iGEM-Team from Norwich developed a biosensor which enables the identification of antimycin-producing strains of Streptomyctes. Therefore they asked us to send them soil samples from Bielefeld, which of course we did. The interesting soil samples of Bielefeld are shown below. We hope that our soil samples helped Norwich.
iGEM-Team UC Davis
The iGEM-Team UC-Davis 2013 wants to develop and implement an
[http://dilbert.cs.ucdavis.edu/Depot/ experimental data depot] for BioBrick characterization data and kindly asked us to collaborate with them. They want to establish an easily accessible and comparative platform to share experimental data about BioBricks and are searching for other teams that are willing to help them establish standardized characterization methods. We tested their promoter characterization protocols on the
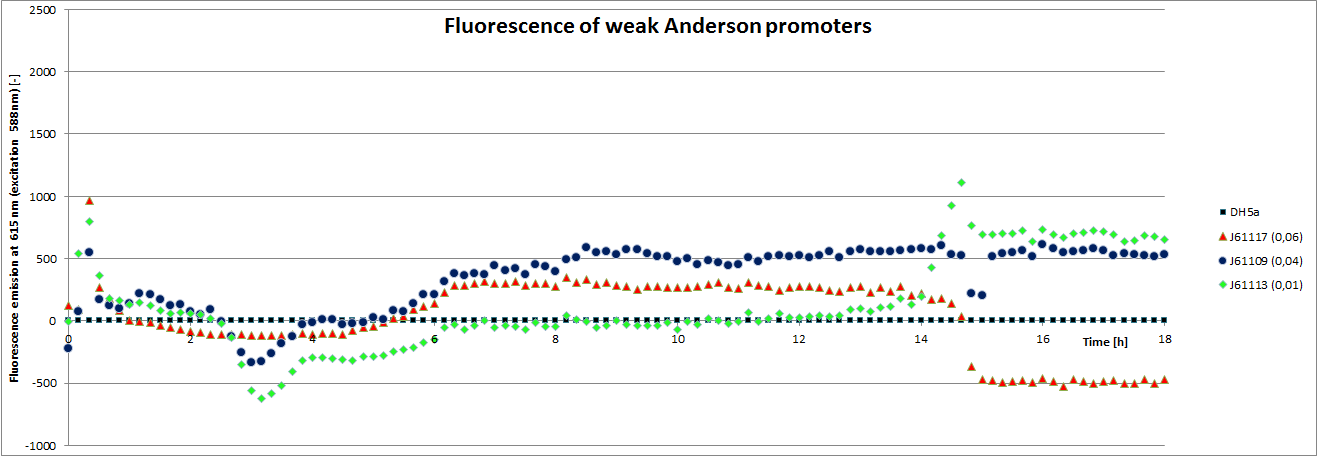
[http://parts.igem.org/Promoters/Catalog/Anderson Anderson promoter collection] J230100-J230119. The plasmid backbone of these BioBricks J61002 is very suitable for promoter insertion between the XbaI and SpeI sites, which places the promoter upstream of a red fluorescent protein (RFP) gene. Depending on the used promoter, the rfp-expression is in- or decreased, while the promoter strength can easily be characterized via fluorescence measurements.
Our team focused mainly on the second half of the collection, which comprises promoters with the below listed strengths.
| BioBrick number | Measured relative promoter strength [-] |
|---|---|
| BBa_J23119 | n/a |
| BBa_J23101 | 0.7 |
| BBa_J2308 | 0.51 |
| BBa_J2309 | 0.04 |
| BBa_J23111 | 0.58 |
| BBa_J23112 | 0.00 |
| BBa_J23113 | 0.01 |
| BBa_J23114 | 0.10 |
| BBa_J23115 | 0.15 |
| BBa_J23116 | 0.16 |
| BBa_J23117 | 0.06 |
| BBa_J23118 | 0.56 |
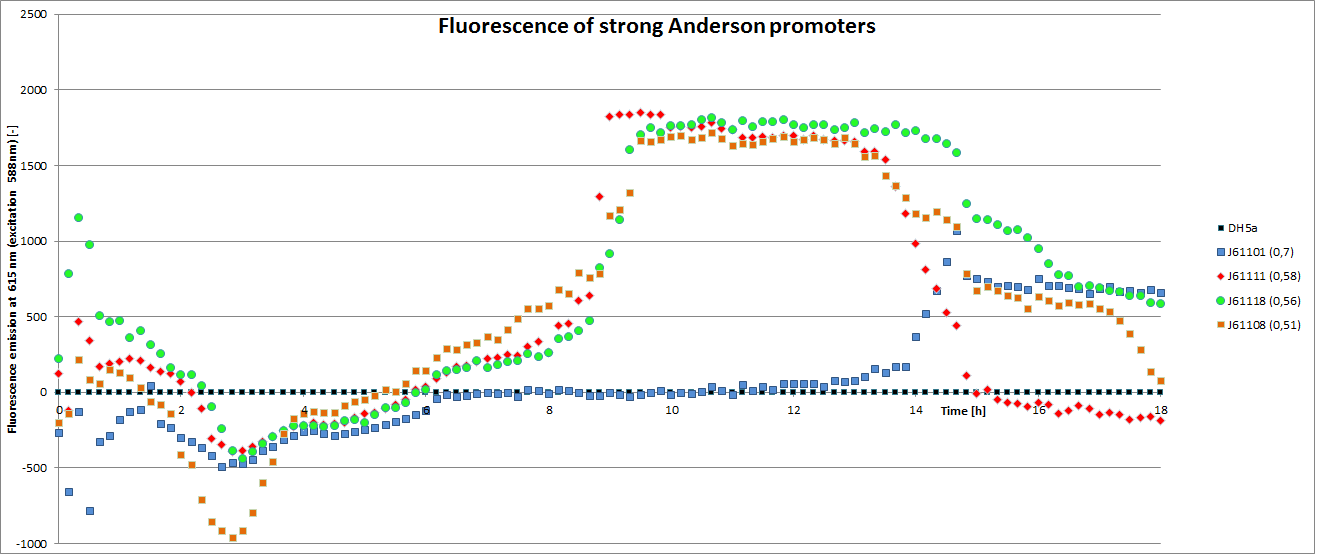
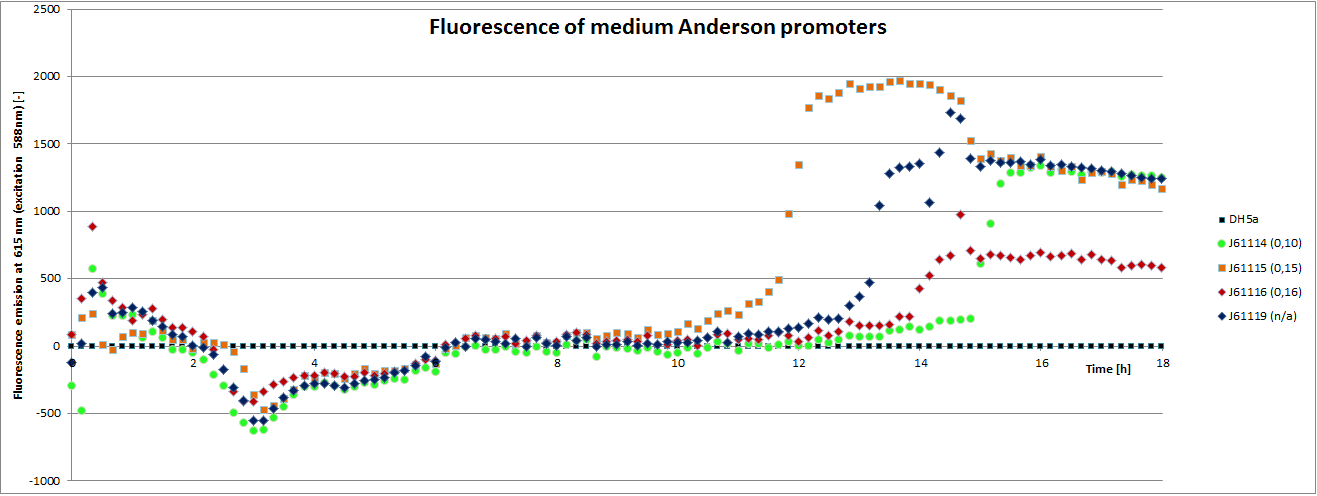
We carried out an extended, 18-hour experiment, monitoring cell density through absorbance measurement and promoter strength by fluorescence emission. The results are shown subsequently.
Please note that we monitored the optical density at 700 nm wavelength, because the UC-Davis team was concerned about interferences of OD600 measurements with the fluorescence measurements.
To determine the relative strength of the used promoters we looked at the fluorescence signal of RFP. With increasing promoter strength, transcription should be enhanced and more RFP will be expressed. This results in a higher fluorescence signal.
The results show very clearly that strains containing a plasmid with a stronger promoter are fluorescing earlier than those with medium and weak promoters. Also, the fluorescence intensity is significantly lower for weak promoters in comparison to the stronger ones. The DH5α-strain with an inherent J23101 plasmid (relative strength 0.7) did surprisingly not fluoresce the strongest. This might be due to a problem with plasmid stability in this strain. Also, the unmeasured J23119 BioBrick seems to hold a promoter of medium strength.
The widely plausible results show, that the protocols for promoter BioBrick characterization work quite well. Comparisons to experimental results from all over the world can be carried out using this method and promise a well characterized and frequently validated parts registry.
This collaboration was a great chance to communicate and discuss data in an international outreach and taught us a lot about other teams work routines and standards. It was a pleasure to contribute in their project to find a way to effectively share complex experimental data and to work out routines for BioBrick characterization
 "
"