Team:Bielefeld-Germany/Biosafety/Biosafety System M
From 2013.igem.org
| Line 96: | Line 96: | ||
Dihydroxyacetone phosphate can be metabolized in the glycolysis pathway, while lactate aldehyde is oxidized to lactate under aerobic conditions and reduced to L-1,2,-propandiol under anaerobic conditions.<br> | Dihydroxyacetone phosphate can be metabolized in the glycolysis pathway, while lactate aldehyde is oxidized to lactate under aerobic conditions and reduced to L-1,2,-propandiol under anaerobic conditions.<br> | ||
The degradation of L-rhamnose can be separated in three steps. In the first step the L-rhamnose is turned into L-rhamnulose by an isomerase (gene ''rhaA''). L-rhamnulose is in turn phosphorylated to L-rhamnulose-1-phosphate by a kinase (gene ''rhaB'') and the sugar phosphate is finally hydrolyzed by an aldolase (gene ''rhaD'') to dihydroxyacetone phosphate and lactate aldehyde ([https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_M#References Baldoma ''et al.'', 1988]).<br> | The degradation of L-rhamnose can be separated in three steps. In the first step the L-rhamnose is turned into L-rhamnulose by an isomerase (gene ''rhaA''). L-rhamnulose is in turn phosphorylated to L-rhamnulose-1-phosphate by a kinase (gene ''rhaB'') and the sugar phosphate is finally hydrolyzed by an aldolase (gene ''rhaD'') to dihydroxyacetone phosphate and lactate aldehyde ([https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_M#References Baldoma ''et al.'', 1988]).<br> | ||
| - | For our Safety-System ''TetOR alive'', the rhamnose promoter P<sub>Rha</sub> is used to control the expression of the repressor TetR and the essential alanine racemase, because this promoter has a very low basal transcription. This is needed to tightly repress the expression of the alanine racemase (''alr'') and thereby take advantage of the double-kill switch. The Tet-System is also characterized by a tight repression, but the induction have to be realized with an antibiotic or anhydrotetracycline. Although the rhamnose promoter P<sub>''Rha''</sub> is characterized by a very low basal transcription it can not be used for the control of the toxic RNase Ba ([https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_M#RNase_Ba_.28Barnase.29 Barnase]), which ist needed for the alanine racemase (''alr''), the other part of the double-kill switch. In conclusion this promoter is the best choice for the first part of the Biosafety-System, as it is tightly repressed in the absence of L-rhamnose, but activated in its presence.</p> | + | For our Safety-System ''TetOR alive'', the rhamnose promoter P<sub>Rha</sub> is used to control the expression of the repressor TetR and the essential alanine racemase, because this promoter has a very low basal transcription. This is needed to tightly repress the expression of the alanine racemase (''alr'') and thereby take advantage of the double-kill switch. The Tet-System is also characterized by a tight repression, but the induction have to be realized with an antibiotic or anhydrotetracycline([https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_M#References Kamionka ''et. al'', 2004]). Although the rhamnose promoter P<sub>''Rha''</sub> is characterized by a very low basal transcription it can not be used for the control of the toxic RNase Ba ([https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_M#RNase_Ba_.28Barnase.29 Barnase]), which ist needed for the alanine racemase (''alr''), the other part of the double-kill switch. In conclusion this promoter is the best choice for the first part of the Biosafety-System, as it is tightly repressed in the absence of L-rhamnose, but activated in its presence.</p> |
<br> | <br> | ||
| Line 104: | Line 104: | ||
[[File:IGEM Bielefeld 2013 biosafety TetR.png|left]][[File:IGEM Bielefeld 2013 biosafety TetO.png|left]] | [[File:IGEM Bielefeld 2013 biosafety TetR.png|left]][[File:IGEM Bielefeld 2013 biosafety TetO.png|left]] | ||
<p align="justify"> | <p align="justify"> | ||
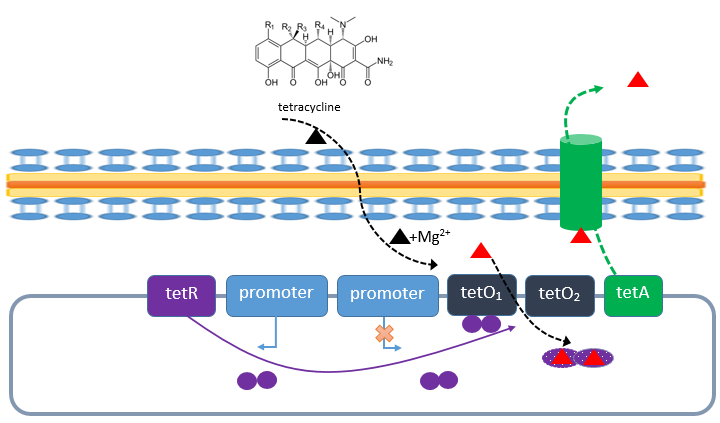
| - | The tetracycline repressor (TetR)/ operator (TetO) is naturally used by ''E. coli'' to inactivate the bacteriolytic effect of the antibiotic tetracycline.In the absence of tetracycline (continous line) TetR is expressed and forms a homodimer and binds with high affinity to the two tetracycline operator sides ''tetO<sub>1</sub>'' and ''tetO<sub>2</sub>''. In the presence of the antibiotic tetracycline, it diffuse through the cell membrane and can inhibit the transpeptidase, which is responsible for the cross-linkage of peptidoglycan. This will result in bacterial cell lysis. When the Tet-System is present the tetracycline uptaken from the environment aggregates with Mg<sup>2+</sup> forming a chelate complex (red triangle). This complex binds to TetR causing a conformation switch in the repressor of the ''tetO'' operator. This causes that the repressor TetR is released from the ''tetO'' operator, resulting in the expression of TetA. TetA is assembled into the cytoplasmic membrane and works as an antiporter by transporting the complex to the outside. This prevent the bacteriolytic effect of the antibiotica. ([https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_M#References | + | The tetracycline repressor (TetR)/ operator (TetO) is naturally used by ''E. coli'' to inactivate the bacteriolytic effect of the antibiotic tetracycline.In the absence of tetracycline (continous line) TetR is expressed and forms a homodimer and binds with high affinity to the two tetracycline operator sides ''tetO<sub>1</sub>'' and ''tetO<sub>2</sub>''. In the presence of the antibiotic tetracycline, it diffuse through the cell membrane and can inhibit the transpeptidase, which is responsible for the cross-linkage of peptidoglycan. This will result in bacterial cell lysis. When the Tet-System is present the tetracycline uptaken from the environment aggregates with Mg<sup>2+</sup> forming a chelate complex (red triangle). This complex binds to TetR causing a conformation switch in the repressor of the ''tetO'' operator. This causes that the repressor TetR is released from the ''tetO'' operator, resulting in the expression of TetA. TetA is assembled into the cytoplasmic membrane and works as an antiporter by transporting the complex to the outside. This prevent the bacteriolytic effect of the antibiotica. ([https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_M#References S Orth ''et. al.'', 2000]). |
</p> | </p> | ||
<br> | <br> | ||
| - | [[File:IGEM Bielefeld 2013 Biosafety tetRnatürlich.png|600px|thumb|center|'''Figure 2:''' Principle of TetR and | + | [[File:IGEM Bielefeld 2013 Biosafety tetRnatürlich.png|600px|thumb|center|'''Figure 2:''' Principle of TetR and ''tetO'' system in tetracycline resistent bacteria.]] |
<br> | <br> | ||
| - | + | For our Biosafety-System ''TetOR alive'' we used this System for the regualtion of the toxic RNase Ba. We used the TetR repressor (<bbpart>BBa_C0040</bbpart>) for the regulation of the ''tetO'' operator (<bbpart>BBa_R0040</bbpart>) containing the toxic RNase Ba. As this System is known for a tight repression and a high activation, it should be optimal for a Biosafety-System. | |
| - | + | ||
| - | + | ||
| Line 217: | Line 215: | ||
*Paddon, C. J. Vasantha, N. and Hartley, R. W. (1989): Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC209718/pdf/jbacter00168-0575.pdf|''Journal of Bacteriology 171: 1185 - 1187]. | *Paddon, C. J. Vasantha, N. and Hartley, R. W. (1989): Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC209718/pdf/jbacter00168-0575.pdf|''Journal of Bacteriology 171: 1185 - 1187]. | ||
| + | |||
| + | *Kamionka, A., Sehnal, M., Scholz, O., Hillen, W. (2004) Independent Regulation of Two Genes in Escherichia coli by Tetracyclines and Tet Repressor Variants [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC421600/pdf/0151-04.pdf|''Journal of Bacteriology 186(13): 4399 – 4401.] | ||
*Voss, Carsten Lindau, Dennis and Flaschel, Erwin (2006): Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use [http://onlinelibrary.wiley.com/doi/10.1021/bp050417e/pdf|''Biotechnology Progress 22: 737 - 744]. | *Voss, Carsten Lindau, Dennis and Flaschel, Erwin (2006): Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use [http://onlinelibrary.wiley.com/doi/10.1021/bp050417e/pdf|''Biotechnology Progress 22: 737 - 744]. | ||
Revision as of 22:20, 28 October 2013
Biosafety System TetOR alive
Overview
The Biosafety-System TetOR alive <bbpart>BBa_K1172915</bbpart> is an improvement of the BioBrick <bbpart>BBa_K914014</bbpart> by replacing the first promoter into the rhamnose promoter PRha, integration of the alanine racemase <bbpart>BBa_K1172901</bbpart> and utilization of the repressor TetR to regulate the transcription of the Barnase behind the tetO promoter. Because this system is known for a tight repression and a fast activation, it is expected that bacteria containing this Biosafety-System are dead or alive...
Genetic Approach
Rhamnose promoter PRha
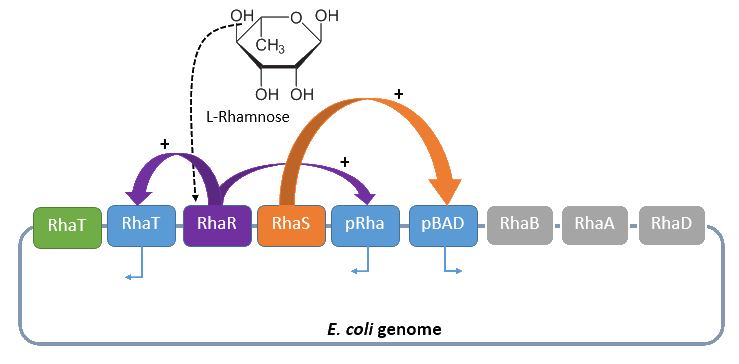
The promoter PRha (<bbpart>BBa_K914003</bbpart>) naturally regulates the catabolism of the hexose L-rhamnose. The advantage to use this promoter is its solely positive regulation. The regulon consists of the gene rhaT for the L-rhamnose transporter and the two operons rhaSR and rhaBAD.
The operon rhaSR encodes the two transcriptional activators RhaS and RhaR, who are responsible for the positive activation of the L-rhamnose catabolism, while the operon rhaBAD encodes the genes for the direct catabolism of L-rhamnose.
When L-rhamnose is present, it acts as an inducer by binding to the regulatory protein RhaR. RhaR regulates its own expression and the expression of the regulatory gene rhaS by repressing or, in the presence of L-rhamnose, activating the operon rhaSR. Normally, the expression level is modest, but it can be enhanced by a higher level of intracellular cAMP, which increases in the absence of glucose. So, in the presence of L-rhamnose and a high concentration of intracellular cAMP, the activator protein RhaR is expressed on higher level, resulting in an activation of the promoter PrhaT for an efficient L-rhamnose uptake and an activation of the operon rhaBAD. The L-rhamnose is than broken down into dihydroxyacetone phosphate and lactate aldehyde by the enzymes encoded by rhaBAD (Wickstrum et al., 2005).
A brief schematic summary of the regulation is shown in Figure 1.
Dihydroxyacetone phosphate can be metabolized in the glycolysis pathway, while lactate aldehyde is oxidized to lactate under aerobic conditions and reduced to L-1,2,-propandiol under anaerobic conditions.
The degradation of L-rhamnose can be separated in three steps. In the first step the L-rhamnose is turned into L-rhamnulose by an isomerase (gene rhaA). L-rhamnulose is in turn phosphorylated to L-rhamnulose-1-phosphate by a kinase (gene rhaB) and the sugar phosphate is finally hydrolyzed by an aldolase (gene rhaD) to dihydroxyacetone phosphate and lactate aldehyde (Baldoma et al., 1988).
For our Safety-System TetOR alive, the rhamnose promoter PRha is used to control the expression of the repressor TetR and the essential alanine racemase, because this promoter has a very low basal transcription. This is needed to tightly repress the expression of the alanine racemase (alr) and thereby take advantage of the double-kill switch. The Tet-System is also characterized by a tight repression, but the induction have to be realized with an antibiotic or anhydrotetracycline(Kamionka et. al, 2004). Although the rhamnose promoter PRha is characterized by a very low basal transcription it can not be used for the control of the toxic RNase Ba (Barnase), which ist needed for the alanine racemase (alr), the other part of the double-kill switch. In conclusion this promoter is the best choice for the first part of the Biosafety-System, as it is tightly repressed in the absence of L-rhamnose, but activated in its presence.
Tetracyclin repressor/operator
The tetracycline repressor (TetR)/ operator (TetO) is naturally used by E. coli to inactivate the bacteriolytic effect of the antibiotic tetracycline.In the absence of tetracycline (continous line) TetR is expressed and forms a homodimer and binds with high affinity to the two tetracycline operator sides tetO1 and tetO2. In the presence of the antibiotic tetracycline, it diffuse through the cell membrane and can inhibit the transpeptidase, which is responsible for the cross-linkage of peptidoglycan. This will result in bacterial cell lysis. When the Tet-System is present the tetracycline uptaken from the environment aggregates with Mg2+ forming a chelate complex (red triangle). This complex binds to TetR causing a conformation switch in the repressor of the tetO operator. This causes that the repressor TetR is released from the tetO operator, resulting in the expression of TetA. TetA is assembled into the cytoplasmic membrane and works as an antiporter by transporting the complex to the outside. This prevent the bacteriolytic effect of the antibiotica. (S Orth et. al., 2000).
For our Biosafety-System TetOR alive we used this System for the regualtion of the toxic RNase Ba. We used the TetR repressor (<bbpart>BBa_C0040</bbpart>) for the regulation of the tetO operator (<bbpart>BBa_R0040</bbpart>) containing the toxic RNase Ba. As this System is known for a tight repression and a high activation, it should be optimal for a Biosafety-System.
Alanine racemase Alr
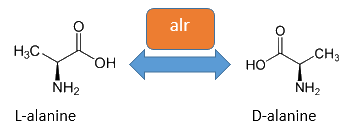
The alanine racemase Alr (EC 5.1.1.1) from the Gram-negative enteric bacteria Escherichia coli is an isomerase, which catalyses the reversible conversion of L-alanine into the enantiomer D-alanine (see Figure 2). For this reaction, the cofactor pyridoxal-5'-phosphate (PLP) is necessary. The constitutively expressed alanine racemase (alr) is naturally responsible for the accumulation of D-alanine. This compound is an essential component of the bacterial cell wall, because it is used for the cross-linkage of peptidoglycan (Walsh, 1989).
The usage of D-alanine instead of a typically L-amino acid prevents cleavage by peptidases. However, a lack of D-alanine causes to a bacteriolytic characteristics. In the absence of D‑alanine dividing cells will lyse rapidly. This fact is used for our Biosafety-Strain, a D-alanine auxotrophic mutant (K-12 ∆alr ∆dadX). The Biosafety-Strain grows only with a plasmid containing the alanine racemase (<bbpart>BBa_K1172901</bbpart>) to complement the D-alanine auxotrophy. Consequently the alanine racemase is essential for bacterial cell division. This approach guarantees a high plasmid stability, which is extremely important when the plasmid contains a toxic gene like the Barnase. In addition this construction provides the possibility for the implementation of a double kill-switch system. Because if the expression of the alanine racemase is repressed and there is no D-alanine supplementation in the medium, cells will not grow.
Terminator
Terminators are essential to terminate the transcription of an operon. In procaryotes two types of terminators exist. The rho-dependent and the rho-independent terminator. Rho-independent terminators are characterized by their stem-loop forming sequence. In general, the terminator-region can be divided into four regions. The first region is GC-rich and constitutes one half of the stem. This region is followed by the loop-region and another GC-rich region that makes up the opposite part of the stem. The terminator closes with a poly uracil region, which destabilizes the binding of the RNA-polymerase. The stem-loop of the terminator causes a distinction of the DNA and the translated RNA. Consequently the binding of the RNA-polymerase is cancelled and the transcription ends after the stem-loop (Carafa et al., 1990).
For our Bioafety-System TetOR alive the terminator is necessary to avoid that the expression of the genes under control of the rhamnose promoter PRha, like the Repressor TetR and the alanine racemase (alr) results in the transcription of the genes behind the tetracylcine operon tetO which contains the toxic Barnase <bbpart>BBa_K1172904</bbpart> and would lead to cell death.
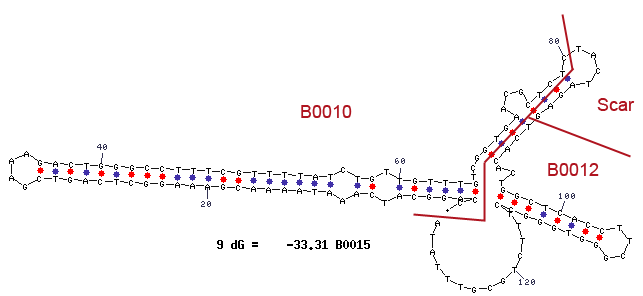
RNase Ba (Barnase)
The Barnase (EC 3.1.27) is a 12 kDa extracellular microbial ribonuclease, which is naturally found in the Gram-positive soil bacteria Bacillus amyloliquefaciens and consists of a single chain of 110 amino acids. The Barnase (RNase Ba) catalyses the cleavage of single stranded RNA, preferentially behind Gs. In the first step of the RNA-degradation a cyclic intermediate is formed by transesterification and afterwards this intermediate is hydrolyzed yielding in a 3'-nucleotide (Mossakowska et al., 1989).
In Bacillus amyloliquefaciens, the activity of the Barnase (RNase Ba) is inhibited intracellular by an inhibitor called barstar. Barstar consists of only 89 amino acids and binds with a high affinity to the toxic Barnase. This prevents the cleavage of the intracellular RNA in the host organism (Paddon et al., 1989). Therefore the Barnase normally acts only outside the cell and is translocated under natural conditions. For the Biosafety-System TetOR alive we modified the enzyme by cloning only the sequence responsible for the cleavage of the RNA, leaving out the N-terminal signal peptide part.
As shown in Figure 6 below, the transcription of the DNA, which encodes the Barnase produces a 474 nt RNA. The translation of the RNA starts about 25 nucleotides downstream from the transcription start and can be divided into two parts. The first part (colored in orange) is translated into a signal peptide at the amino-terminus of the Barnase coding RNA. This part is responsible for the extracellular translocation of the RNase Ba, while the peptide sequence for the active Barnase starts 142 nucleotides downstream from the transcription start (colored in red).
For the Biosafety-System TetOR alive, we only used the part (<bbpart>BBa_K1172904</bbpart>) of the Barnase encoding the catalytic domain without the extracellular translocation signal of the toxic gene product. Translation of the barnase gene leads to rapid cell death if the expression of the Barnase is not repressed by the repressor TetR of our Biosafety-System.
Biosafety system TetOR alive
Combining the genes described above with the Biosafety-Strain K-12 ∆alr ∆dadX results in a powerful device, allowing us to control the bacterial cell division. The control of the bacterial growth is possible either active or passive. Active by inducing the tetO operon with tetracycline and passive by the induction of L-rhamnose. The passive control makes it possible to control the bacterial cell division in a defined closed environment, like the MFC, by continuously adding L-rhamnose to the medium. As shown in Figure 7 below, this leads to an expression of the essential alanine racemase (alr) and the TetR repressor, so that the expression of the RNase Ba is repressed.
In the event that bacteria exit the defined environment of the MFC or L-rhamnose is not added to the medium any more, both the expression of the alanine Racemase (Alr) and the TetR repressor decrease, so that the expression of the toxic RNase Ba (Barnase) begins. The cleavage of the intracellular RNA by the Barnase and the lack of synthesized D-alanine, caused by the repressed alanine racemase inhibit the cell division. Through this it can be secured that the bacteria can only grow in the defined area or the device of choice respectively.

</p>
Results
Characterization of the tetracycline tetO operator
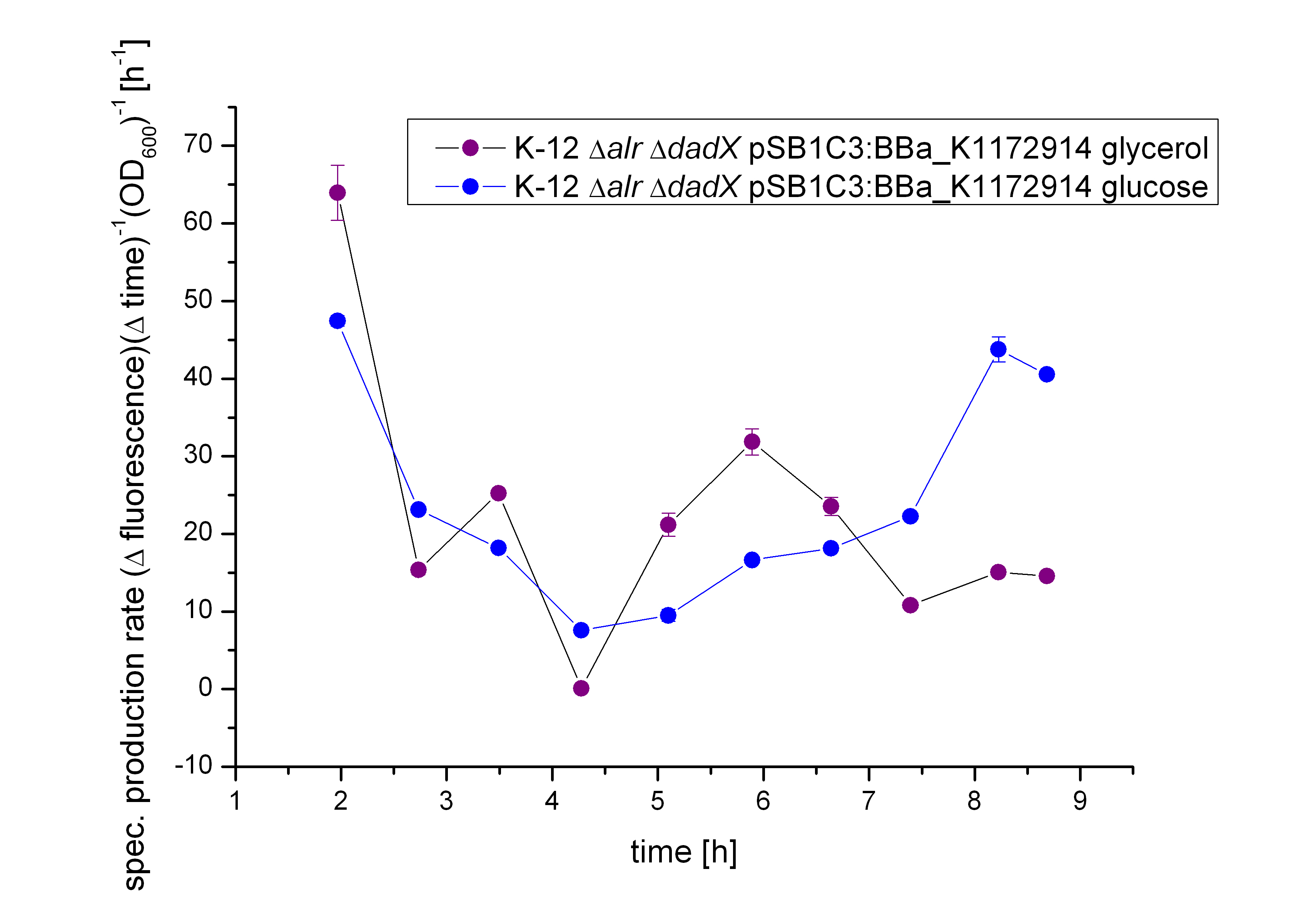
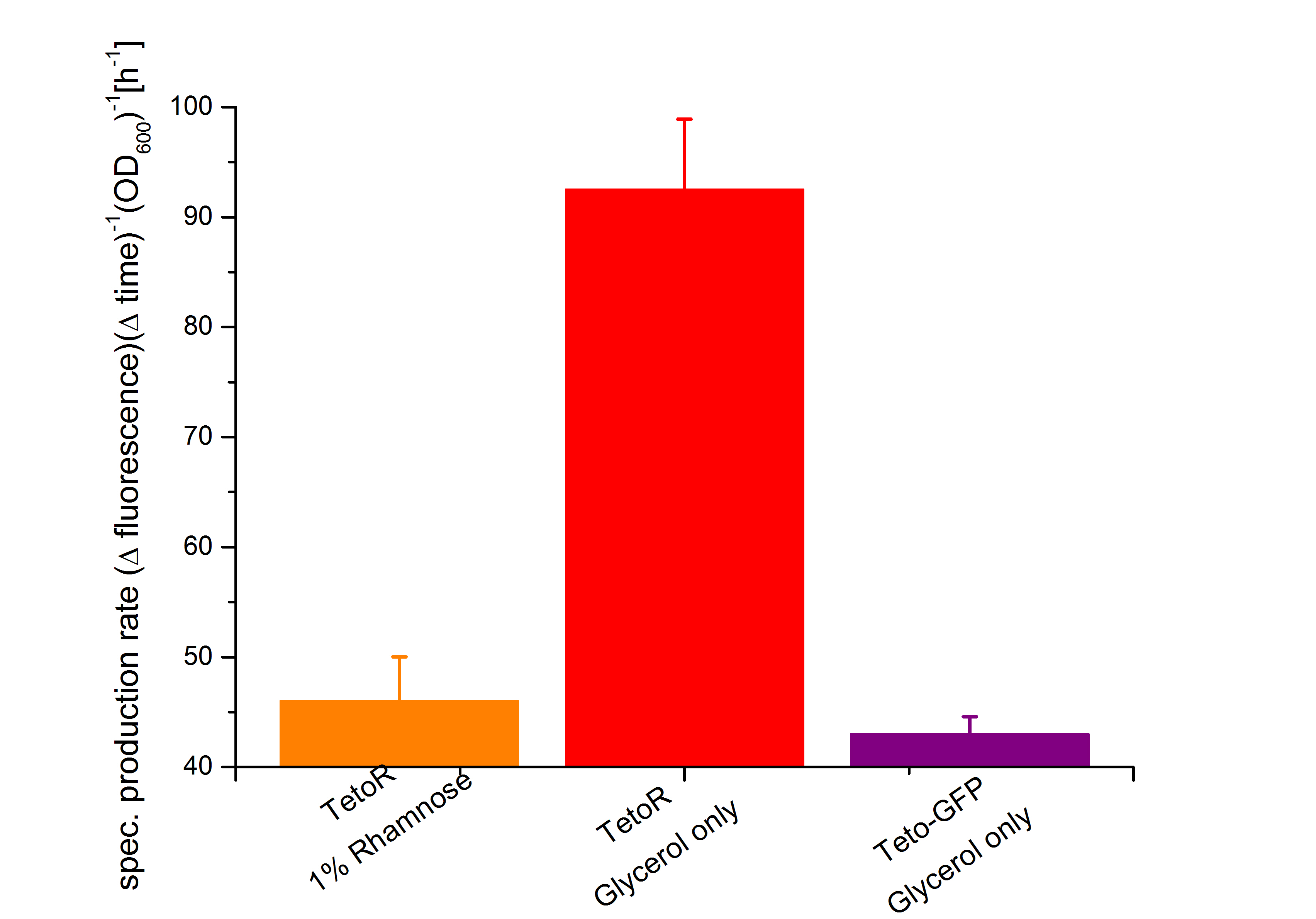
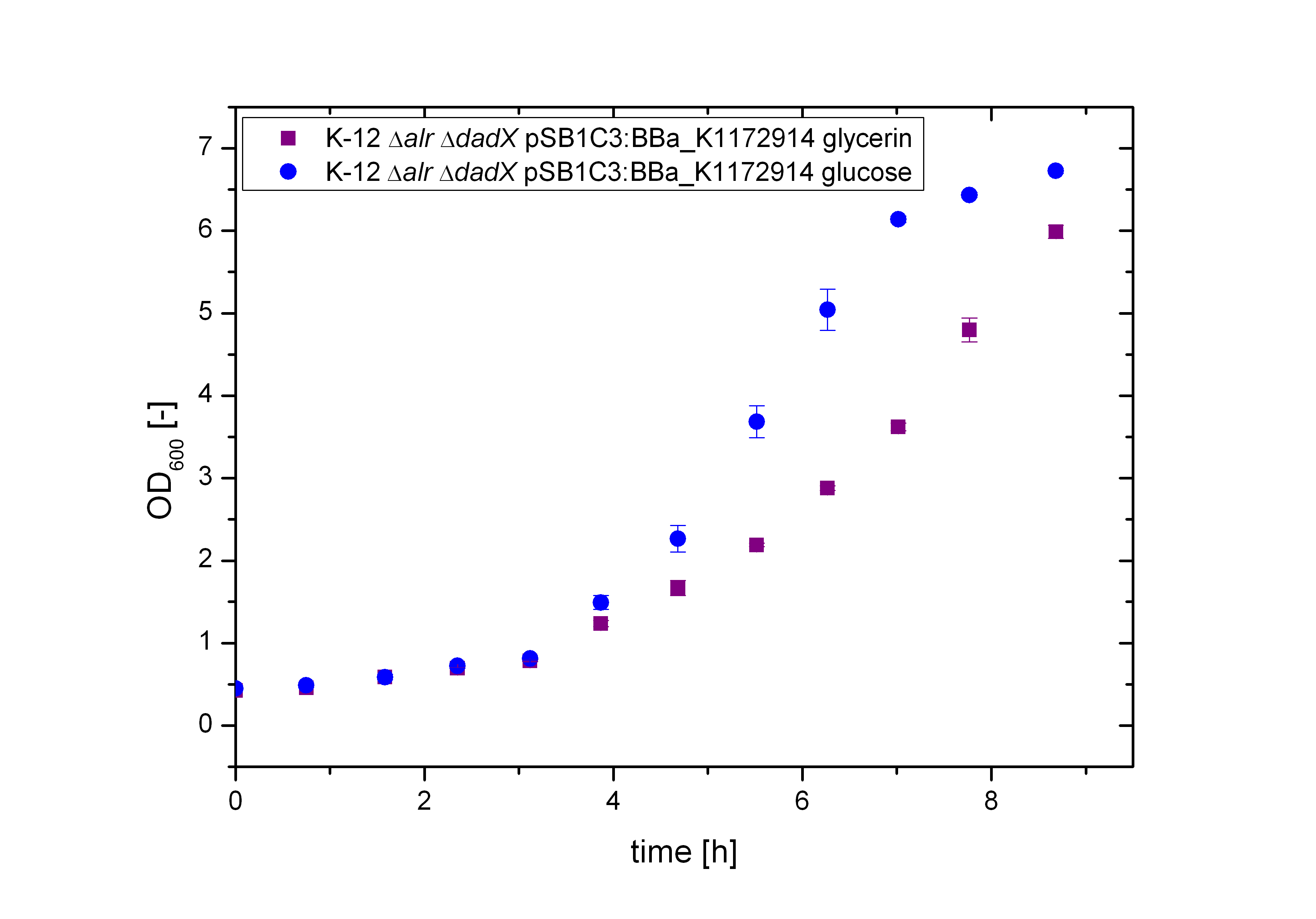
First of all the bacterial growth under the pressure of the unrepressed tetracycline operator TetO was investigated on different carbon source. Therefore the cultivation on M9 minimal media with glycerin or glucose was characterized. To identify the transcription rate of the unexpressed tetracycline operator TetO the expression of the green fluorescence protein GFP BBa_E0040 behind the tetracycline operator TetO was used.
As shown in figure 10 below, the bacteria adapted better on glucose then on glycerol. As glucose is the more powerful energy source, because it posses more carbon atoms than glycerol these result was expected before. So more interesting are the fluorescence measurement shown in the figure 11. As it can be seen also the fluorescence depends on the carbon source, but not as strong as it can be seen by the arabinose promoter pBAD. The Biosafety-Strain K-12 ∆alr ∆dadX shows a little higher expression of GFP with glycerol than with glucose. This difference between the two curves can be seen as insignificant.
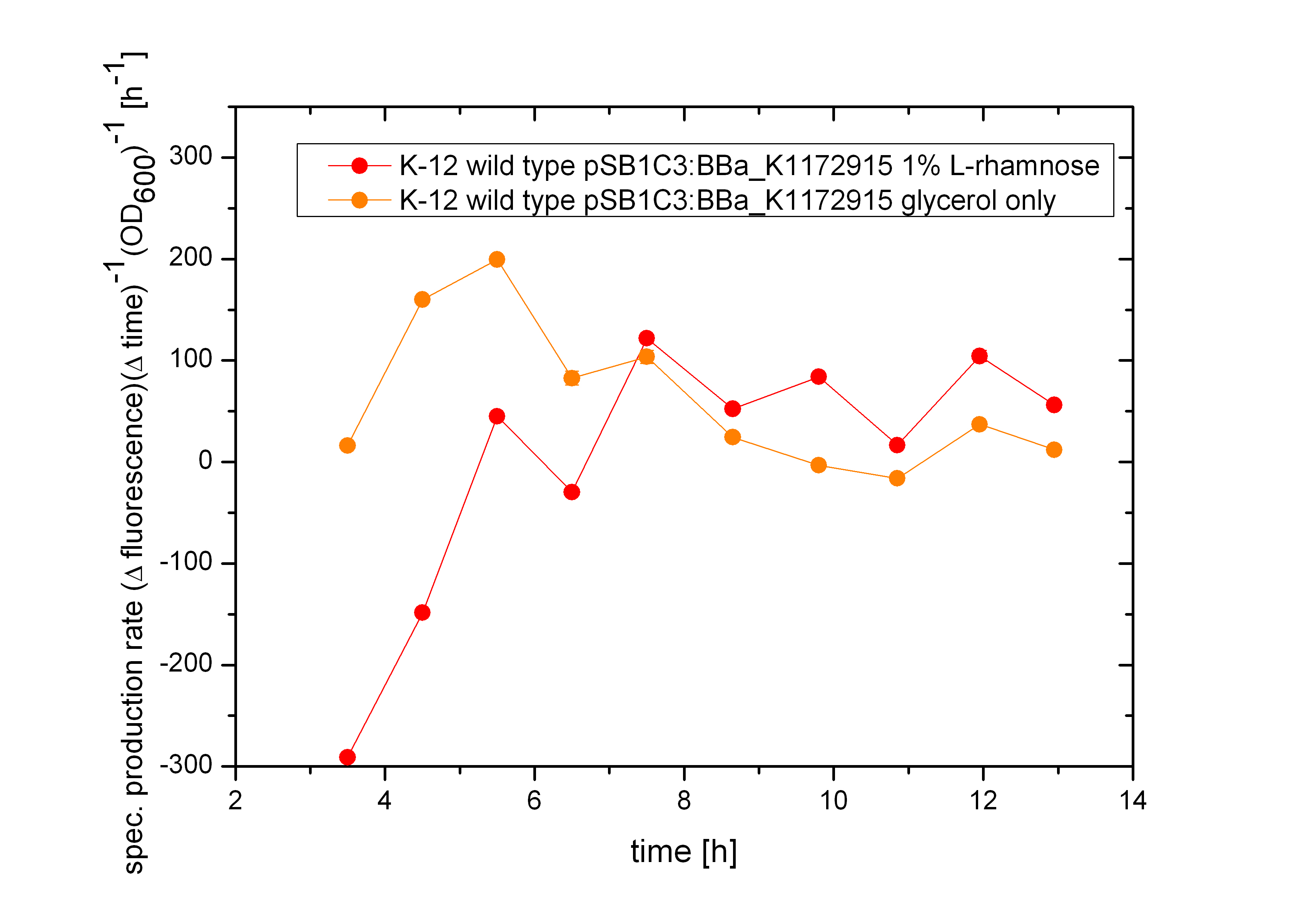
It can be seen that the expression of GFP between the two cultivations doesn’t have that difference like the systems Aractive or the lac of growth have. As the production rate by using glucose as carbon source is nearly the same like the cultivation with glycerol. As the specific production rate was calculated between every single measurement point the curve is not smoothed and so the fluctuations have to be ignored, as they do not stand for are real fluctuations in the transcription in the expression of GFP. They are caused by the growth curve and the fluorescence curve. And as they are not ideal there exists the fluctuations. But this graph shows that there is an insignificant difference between the two carbon sources.
Characterization of the Biosafety-System TetOR alive
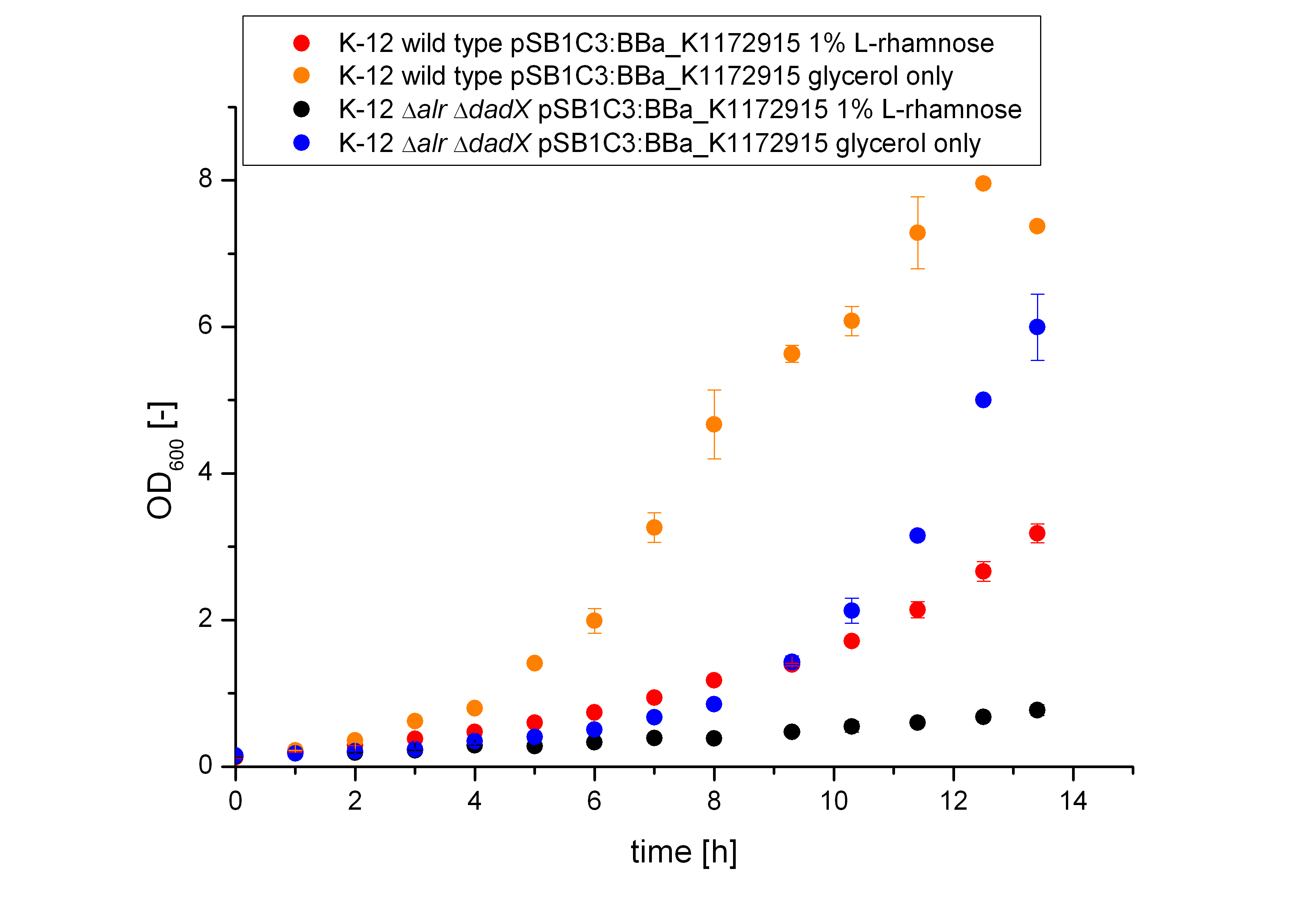
The Biosafety-System TetOR alive was characterized on M9 minimal medium using glycerol as carbon source. As for the characterization of the pure tetracycline promoter above, the bacterial growth and the fluorescence of GFP <bbpart>BBa_E0040</bbpart> was measured. Therefore, the wild type and the Biosafety-Strain E. coli K-12 ∆alr ∆dadX containing the Biosafety-Plasmid <bbpart>BBa_K1172912</bbpart> were cultivated once with the induction of 1% L-rhamnose and once only on glycerol.
It becomes obvious (Figure 13) that the bacteria, induced with 1 % L-rhamnose (red and black curve) grow significantly slower than on pure glycerol (yellow and blue curve). This is attributed to the high metabolic burden encountered by the induced bacteria. The expression of the repressor AraC and the alanine racemase (Alr) simultaneously causes a high metabolic stress on the cells, so that they grow slower than the uninduced cells, who expresses only GFP. Additionally the tetracycline promoter is tightly regulated, so that the expression even with a small amount of the repressor AraC is not that high and therefore not as stressful as the induced expression of the AraC repressor and the alanine racemase (Alr).
Comparing the bacterial growth with the fluorescence in figure 14 it can be seen that the fluorescence of the Biosafety-Strain can not be evaluate because of the long duration of the lac-phase.
From the figure above it can not be seen if the expression of the repressor TetR does effect the transcription of GFP or not. The slower growth of the bacteria is a first indication that the repressor tetR and the alanine racemase are highly expressed. As the specific production rate was calculated between every single measurement point the curve is not smoothed and so the fluctuations have to be ignored, as they do not stand for are real fluctuations in the transcription in the expression of GFP. They are caused by the growth curve and the fluorescence curve. And this measured curves are not ideal the calculation of the specific production rate causes the fluctuations. But it can be seen that the production of GFP differs and tends to be lower, when the bacteria are induced with 1% L-Rhamnose. So the Biosafety-System tetOR alive works.
Conclusions
References
- Saenger W. et al. (2000):Der Tetracyclin-Repressor – das Musterbeispiel für einen biologischen Schalter, In:[http://onlinelibrary.wiley.com/doi/10.1002/1521-3757%2820000616%29112:12%3C2122::AID-ANGE2122%3E3.0.CO;2-8/abstract Angewandte Chemie Volume 112, Issue 12, pages 2122–2133]
- Mossakowska, Danuta E. et al. (1989): Kinetic Characterization of the Recombinant Ribonuclease from Bacillus amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis [http://pubs.acs.org/doi/pdf/10.1021/bi00435a033 Biochemistry 28: 3843 - 3850].
- Paddon, C. J. Vasantha, N. and Hartley, R. W. (1989): Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC209718/pdf/jbacter00168-0575.pdf|Journal of Bacteriology 171: 1185 - 1187].
- Kamionka, A., Sehnal, M., Scholz, O., Hillen, W. (2004) Independent Regulation of Two Genes in Escherichia coli by Tetracyclines and Tet Repressor Variants [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC421600/pdf/0151-04.pdf|Journal of Bacteriology 186(13): 4399 – 4401.]
- Voss, Carsten Lindau, Dennis and Flaschel, Erwin (2006): Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use [http://onlinelibrary.wiley.com/doi/10.1021/bp050417e/pdf|Biotechnology Progress 22: 737 - 744].
- Orth P. et al. (2000): Structual basis of gene regulation by the tetracycline inducible Tet repressor-operator system. In: [http://life.nthu.edu.tw/~b871641/tetrepressor.pdf nature structural biology, volume 7 number 3].
 "
"