Team:Bielefeld-Germany/Labjournal/July
From 2013.igem.org
July
Milestones
- Poster presentation at the congress ‘[http://www.biotechnologie2020plus.de Next generation of biotechnological processes 2020+]’ in Berlin.
- Sponsoring acquisition is nearly finished.
- Problems with cloning delay the progress in the laboratory.
- We have published our [http://ekvv.uni-bielefeld.de/blog/uniaktuell/entry/mit_bakterien_batterie_strom_erzeugen first press release] with a great response. The press release was picked up worldwide.
Week 9
Organization
- We’ve presented us at the congress ‘[http://www.biotechnologie2020plus.de Next generation of biotechnological processes 2020+]’ in Berlin at June 27th, 2013. We met the expert (Dr. Falk Harnisch) on the topic of MFC and decided to participate in the 'Day of Synthetic Biology in Germany'.
MFC
Cytochromes
- Design and order of new primers
- The mtrCAB fragment has two illegal PstI restriction sites at 1793 bp and 2078 bp, so specific primers to remove them had to be designed. One base in each restriction site was replaced by respective primer overlaps without affecting the coding triplet. The three different fragments will be ligated back together and into the pSB1C3 shipping plasmid via Gibson Assembly.
- mtrCAB_Frag1_rev (30 nt): GACCTGTTTTATCTGTTGCAGCGGCAGTAT
- mtrCAB_Frag2_fwd (30 nt): ATACTGCCGCTGCAACAGATAAAACAGGTC
- mtrCAB_Frag2_rev (30 nt): GGTGTGACGACTAATGCCATAATTGCAGAC
- mtrCAB_Frag3_fwd (30 nt): GTCTGCAATTATGGCATTAGTCGTCACACC
Nanowires
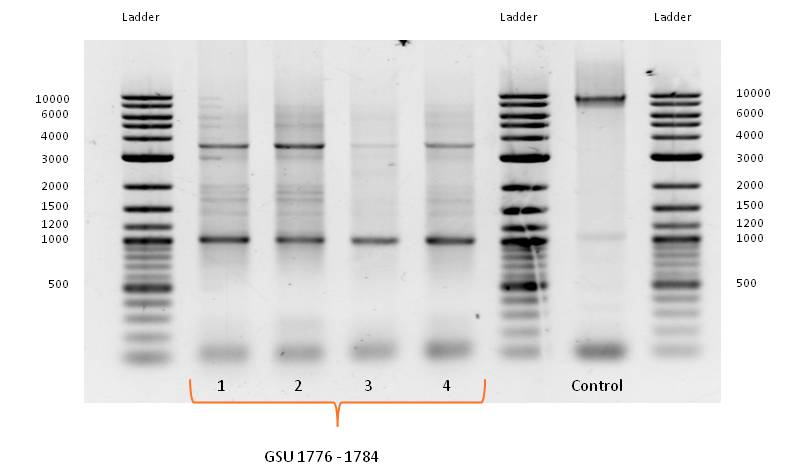
- Primer design for isolation of gene-cluster GSU 1776-1784 from Geobacter sulfurreducens, containing overlaps for BioBrick Prefix and Suffix:
- Forward GSU 1776-1784 (41):
GAATTCGCGGCCGCTTCTAGATGCTGAAACGTTTTCGTAAC - Reverse GSU 1776-1784 (44):
CTGCAGCGGCCGCTACTAGTATCATCCTACAGTGCCGGCAATTT
- Forward GSU 1776-1784 (41):
- Successful genome isolation from Geobacter sulfurreducens strain, using the GenEluteTM Bacterial Genomic DNA-Kit to prevent vigorous fragmentation of the DNA, which arises out of the standard genome isolation method with a ribolyse-step.
- Successful PCR with forward and reverse primer GSU Promoter 1496-1505 and GSU 1496-1505 on the appropriate gene-cluster of Geobacter sulfurreducens, using the PCR Master Mix, described in Table 3 (June) (nanowires), with adjusted template volume and the standard PCR program for amplification of Geobacter sulfurreducens gene cluster (Table 4, June).
- GSU Promoter 1496-1505 and GSU 1496-1505 PCR products were isolated and purified by gel extraction.
Week 10
Organization
- We can present new sponsors of iGEM-Team Bielefeld: Stockmeier, Baxter, Promega, BIO.NRW and AppliChem will support our team.
MFC
Mediators
- Glycerol dehydrogenase
- Further work on gldA BioBrick is needed: Plasmid restriction analysis and colony PCR could not give sufficient results.
- Successful PCR (standard Phusion PCR) on the <bbpart>BBa_J04450</bbpart> BioBrick plasmid using forward primer pSB1C3 and reverse primer pSB1C3 for later Gibson Assembly.
- Gibson Assembly with gldA PCR product and pSB1C3 PCR product.
- Screening of colonies from Gibson Assembly with colony PCR and plasmid restriction analysis shows re-ligated pSB1C3, as already seen while using NEB Assembly Kit.
Porines
- We will try to use Gibson Assembly for cloning oprF into pSB1C3.
- Gibson Assembly with oprF PCR product and pSB1C3 PCR product. Screening of colonies from Gibson Assembly with colony PCR and plasmid restriction analysis shows re-ligated pSB1C3, as already seen while using NEB Assembly Kit.
Nanowires
- PCR with forward and reverse primer GSU 1776-1784 on the appropriate gene-cluster of Geobacter sulfurreducens strain, using the PCR Master Mix, described in Table 3 (June) with adjusted template volume and the standard PCR program for amplification of Geobacter sulfurreducens gene cluster (Table 4, June) did not work as expected.
- Bands of several sizes, but no band at the expected size of 9000 bp, the length of GSU 1776-1784. Because of the correct band for the control PCR on the gene cluster GSU Promoter 1496-1505 a problem with the PCR program can be eliminated.
Week 11
Organization
- We’ve presented us at the congress '[http://www.bio.nrw.de/studentconvention BioNRW pHD Student Convention]' in Düsseldorf at July 13th, 2013 with a short presentation about iGEM and our project.
- We proudly present our [http://ekvv.uni-bielefeld.de/blog/uniaktuell/entry/mit_bakterien_batterie_strom_erzeugen first press release.] It was amazing to see how often our press release was picked up.
MFC
Mediators
- Glycerol dehydrogenase
- Repeating Gibson Assembly shows every time a re-ligated pSB1C3, a Gibson overlap of Prefix and Suffix with 20 bp seems to be too short, furthermore Prefix and Suffix have too many homologous regions.
Porines
- Repeating Gibson Assembly shows every time a re-ligated pSB1C3, a Gibson overlap of Prefix and Suffix with 20 bp seems to be too short, furthermore Prefix and Suffix have too many homologous regions.
Nanowires
- Gibson Assembly with GSU 1491-1495, GSU 1496-1505 and GSU Promoter 1496-1505 PCR product as one fragment and the pSB1C3 PCR product, using the primer with Prefix and Suffix specific overlaps, as the second DNA fragment.
- Screening of colonies from Gibson Assembly by plasmid restriction analysis shows fragments of 2000 bp, which corresponds to re-ligated pSB1C3.
- PCR with forward and reverse primer GSU 1776-1784 on the appropriate gene-cluster of Geobacter sulfurreducens strain, using the PCR Master Mix, described in Table 3 (June) with adjusted template volume was repeated with the standard PCR program for amplification of Geobacter sulfurreducens gene cluster (Table 4, June) modified by a gradient of different annealing temperatures in a range of 45°C to 65°C.
- No bands at the expected size of 9000 bp.
Week 12
Organization
- Dr. Falk Harnisch will have a presentation at CeBiTec colloquium as an expert for our team.
- Preparing experiments for the ‘Day of Synthetic Biology’. Our ideas are experiments like DNA isolation from fruit and vegetables, pipetting of bright colors, chromatography with markers, a potato battery and microscopy.
- The TV station [http://www1.wdr.de/mediathek/video/sendungen/lokalzeit/lokalzeit-owl WDR] would like to contribute with us on our topic.
MFC
Mediators
- Glycerol dehydrogenase
- New primer design for pSB1C3 with longer overlaps individually for each part in order to stop the problem of re-ligated pSB1C3:
- Forward primer pSB1C3 gldA (43 bp): TCCTGCAAGAGTGGGAATAATACTAGTAGCGGCCGCTGCAGTC
- Reverse primer pSB1C3 gldA (43 bp): GATTGAATAATGCGGTCCATCTAGAAGCGGCCGCGAATTCCAG
Cytochromes
- Amplification of Fragment1 with Phusion polymerase
- Size: 1833 bp
- Gradient: 54°C - 71°C over 8 steps
- Primer: 'mtrC'_fwd & mtr_Frag1_rev
- Template: S. oneidensis PCR Template from genomic DNA
- Notes: Annealing temperature has no significant impact on the PCR
- Amplification of Fragment2 with Phusion polymerase
- Size: 321 bp
- Primer: mtr_Frag2_fwd & mtr_Frag2_rev
- Template: S. oneidensis PCR Template from genomic DNA
- Notes: Annealing temperature has no significant impact on the PCR
- Amplification of Fragment3 with Phusion polymerase
- Size: 3057 bp
- Gradient: 54°C - 71°C over 8 steps
- Primer: mtr_Frag3_fwd & mtrB_rev
- Template: S. oneidensis PCR Template from genomic DNA
- Notes:
- Amplification of ccmAH cluster
- Size:6311 bp
- Program: PhusionPCR
- Gradient: 51.5°C - 68°C over 8 steps
- Primer: ccmAH_fwd & ccmAH_rev
- Template: E. coli PCR Template from genomic DNA
- Notes: A high annealing temperature of 68°C or more increases yield as well as reduces unspecific bands and is therefore recommended.
- Gel extraction and cleanup of Fragment 1, 2, 3 and ccmAH
- Elution buffer was preheated to 50°C
- We eluted each column twice, with 20 ul each and combined it afterwards
- Measurement of nucleic acid concentration via NanoDrop
- Fragment1: 4-2607-304: 10.5 ng/ul
- Fragment2: 4-2707-003: 44.6 ng/ul
- Fragment2: 4-2707-004: 13.5 ng/ul
- Fragment3: 4-2607-301: 11.1 ng/ul
- Fragment3: 4-2607-302: 8.9 ng/ul
- Fragment3: 4-2607-303: 9.2 ng/ul
- ccmAH: 4-2807-302: 11.0 ng/ul
- ccmAH: 4-2807-303: 11.5 ng/ul
Porines
- New primer design for pSB1C3 with longer overlaps individually for OprF part in order to stop the problem of re-ligated pSB1C3:
- Forward primer pSB1C3 oprF (43 bp): TTGAAGCCCAAGCTAAGTAATACTAGTAGCGGCCGCTGCAGTC
- Reverse primer pSB1C3 oprF (43 bp): AAGGTGTTTTTCAGTTTCATCTAGAAGCGGCCGCGAATTCCAG
Nanowires
- Multiple Gibson Assembly retries with different molar ratios of the two fragments results every time in re-ligated pSB1C3 plasmids, so that the used Gibson overlap of Prefix and Suffix is not useable.
- Design of new primers for pSB1C3 amplification with longer sequence-specific overlaps for every part:
- Forward pSB1C3 GSU 1491-1505 / GSU 1496-1505 / GSU Promoter 1496-1505 (43 bp):
GGGACGAGGTTCGTTTATGATACTAGTAGCGGCCGCTGCAGTC - Reverse pSB1C3 GSU 1496-1505 (43 bp):
GTATGGGGGTAATTGGCCAACTCTAGAAGCGGCCGCGAATTCC - Forward pSB1C3 GSU 1491-1495 (43 bp):
TTGGCATGGATGAGGAGTGATACTAGTAGCGGCCGCTGCAGTC - Reverse pSB1C3 GSU 1491-1495 / GSU 1491-1505 (43 bp):
CCCAGTCTGCTAGCCTGCATCTAGAAGCGGCCGCGAATTCCAG - Reverse pSB1C3 GSU Promoter 1496-1505 (43 bp):
ACTCGGTGACGGATCCTATCCTCTAGAAGCGGCCGCGAATTCC
- Forward pSB1C3 GSU 1491-1505 / GSU 1496-1505 / GSU Promoter 1496-1505 (43 bp):
Week 13
Organization
- Spontaneous visit of [http://www.radiobielefeld.de Radio Bielefeld] in our laboratory. In addition to a radio interview a [http://www.radiobielefeld.de/programm/bei-uns-im-programm/studenten-entwickeln-biobatterie.html short video clip] was filmed.
MFC
Mediators
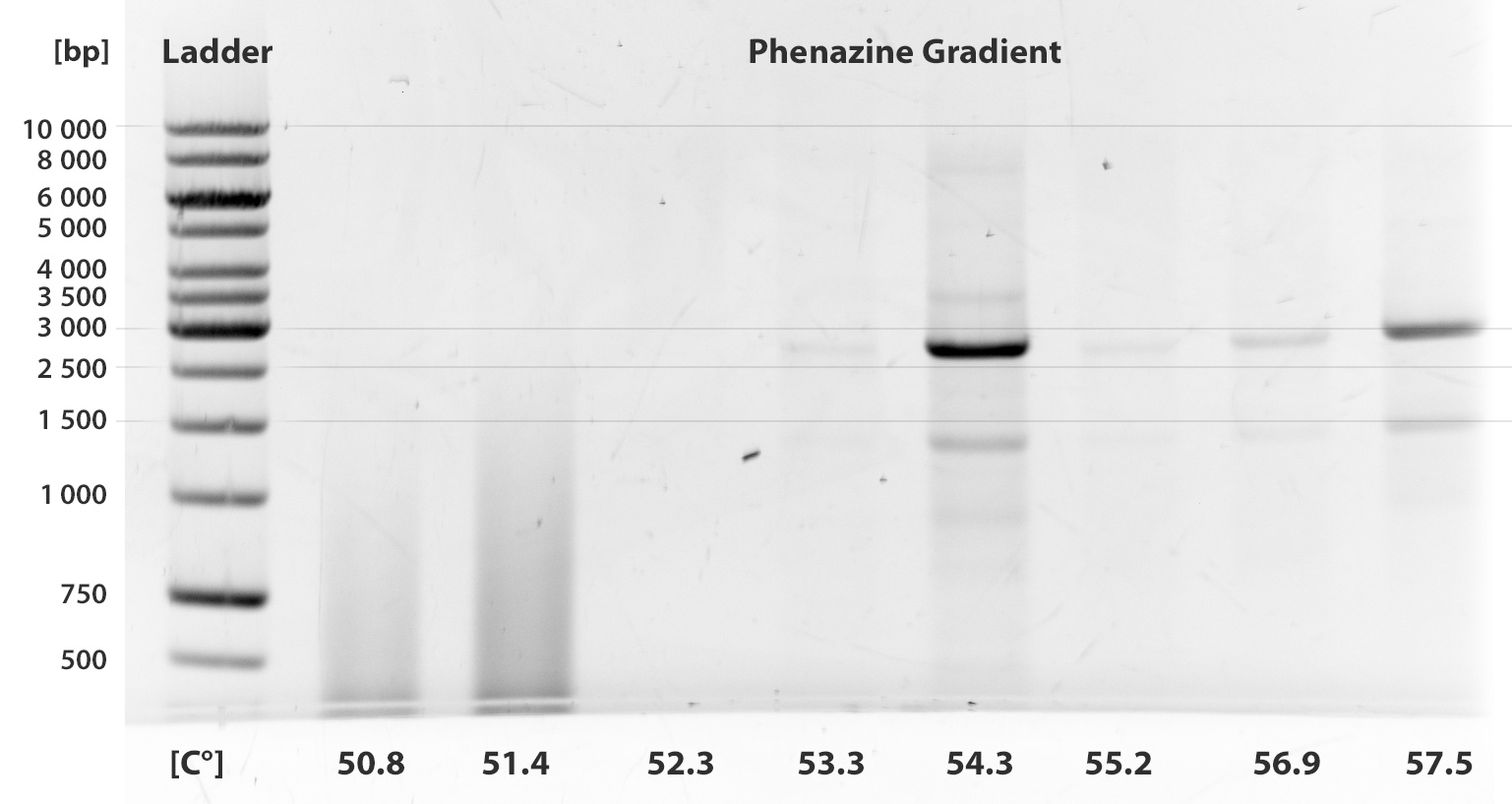
- Phenazine
Almost ten PCRs were run up to this time. Unfortunately, none of them was successful. We show only two gel pictures out of many, where two different gradients were taken.
We have tried to add betaine as a stabilizing agent, work with [http://www.millipore.com/publications.nsf/a73664f9f981af8c852569b9005b4eee/31140b3dbc2b4146852579d8005e9c72/$FILE/PR3366EN00.pdf KOD Hot Start polymerase] according to the manual. We tried different extention times and annealing in different temperatures.
The fragment PhzABCDEFGH contains 8 illegal restriction sites and is 8505 bp long.
After trying all strategies, we decided to abandon this sub-project and start with another endogenous mediator, riboflavin (see Labjournal August)
- Recipe PCR
- 2x KOD Xtreme™ PCR Buffer: 25 µL
- dNTPs: 1.0 µL
- Primer1: 1.0 µL
- Primer2: 1.0 µL
- Template DNA: 1.5 µL
- DMSO: 1.5 µL
- Betaine: 12.5 µL
- KOD Hot Start Polymerase: 0.5 µL
- Dest. H2O: 6 µL
- Total volume: 50 µL
- Protocol PCR Two-step Cycling
- Polymerase Activation 94°C for 2 min
- Denaturation 98°C for 10 s
- Annealing gradient 50.8°C-70.2°C
- Extension 72°C for 270 s/320 s
As is clear from the gels, we were not able to amplify the fragment and have closed this sub-project.
Biosafety
- Amplification of different fragments:
- Recipe:
- PhusionBuffer: 5.5 µL
- dNTPs: 1 µL
- Primer1: 1 µL
- Primer2: 1 µL
- Template: 1.5 µL
- DMSO: 0.5 µL
- Phusion: 0.5 µL
- Dest. H2O: 39 µL
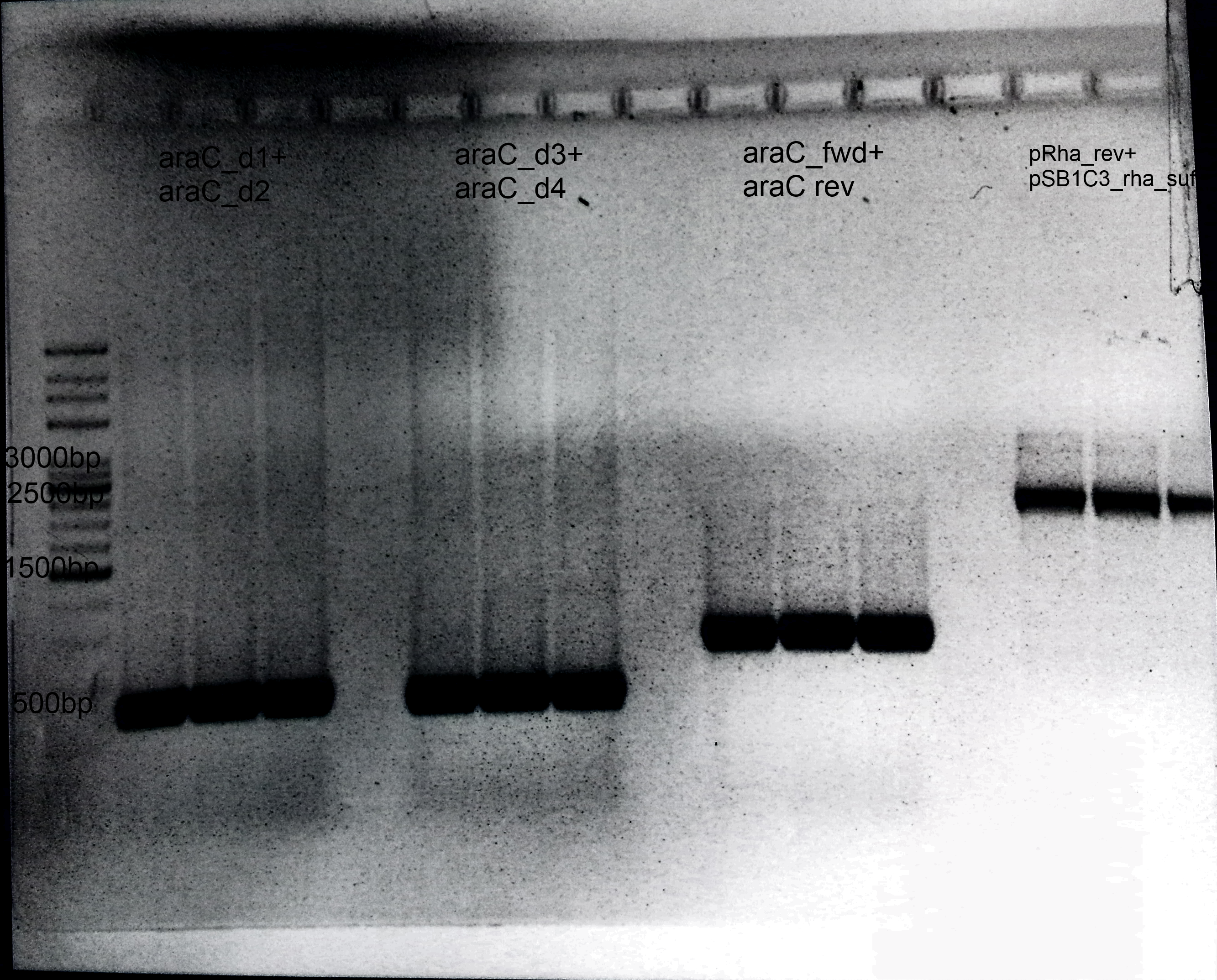
- 1.araC:
- Primer: araC_d1+ araC_d2
- Annealing: 62°C
- Extension: 1 min
- Template: E.coli Genome
- Size: 600 bp
- 2.Deletion of araC:
- Primer: araC_d3+ araC_d4
- Annealing= 62°C
- Extension: 1 min
- Template: E.coli Genome
- Size: 600 bp
- 3.Amplification of araC into our Biosafety system:
- Primer: araC_fwd+ araC rev
- Annealing= 62°C
- Extension: 1 min
- Template: BBa_I13458
- Size: 900 bp
- 4.Preparation of pSB1C3:
- Primer: pRha_rev+ pSB1C3_rha_suf
- Annealing: 62°C
- Extension: 1.30 min
- Template: BBa_I13541
- Size: 3 kb
- 5.Preparation of pSB1C3:
- Primer: pSB1C3_alr_fwd+ pSB1C3_alr_rev
- Annealing: 62°C
- Extension: 1 min
- Template: E.coli Genome
- Size: 1 kb
- 6.Preparation of pSB1C3:
- Primer: pSB1C3_plac_alr+ pSB1C3_alr_rev
- Annealing: 62°C
- Extension: 1 min
- Template: E.coli Genome
- Size: 1 kb
- 7.Preparation of pSB1C3:
- Primer: pSB1C3_alr_pre+ pSB1C3_alr_suf
- Annealing: 62°C
- Extension: 1.30 min
- Template: pSB1C3
- Size: 2 kb
- 8.Preparation of pSB1C3:
- Primer: pSB1C3_plac_pre+ pSB1C3_alr_suf
- Annealing: 62°C
- Extension: 1.30min
- Template: pSB1C3
- Size: 2 kb
- Analysis by agarose gel
- Result:failed
- New PCR:
- Program: 06 phy_cyt mit 62°C Annealing
- 10 µL 5xHF-Buffer
- 4 µL dNTPs
- 0.5 µL Primer1
- 0.5 µL Primer2
- 1 µL Template
- 1.5 µL DMSO
- 1 µL Enzyme
- 31.5 µL dest. H2O
- Analysis by agarose gel:
- PCR-Purification:
- BS1a (9-38-451) 113.6 ng/µL
- BS1b (9-38-452) 75.1 ng/µL
- BS2a (9-38-453) 85.5 ng/µL
- BS3a (9-38-454) 107.6 ng/µL
- BS4a (9-38-455) 14.4 ng/µL
- BS5a (9-38-456) 44.5 ng/µL
- BS5b (9-38-457) 49.0 ng/µL
- BS6a (9-38-458) 63.6 ng/µL
- BS7a (9-38-459) 6.9 ng/µL
- BS8a (9-38-460) 8.6 ng/µL
- BS1a (9-38-451) 113.6 ng/µL
- Successful PCR on the BBa_J04450 BioBrick plasmid using part specific forward and reverse primer (Week 12) for later Gibson Assembly. Isolation of the pSB1C3 PCR product by agarose gel electrophoresis and gel extraction.
- Gibson Assembly, using the different new pSB1C3 PCR products and the according gene-cluster PCR products.
 "
"