Team:Bielefeld-Germany/Project/GldA
From 2013.igem.org
GldA
Glycerol dehydrogenase GldA - Overview
The Gene GldA encodes glycerol dehydrogenase from Escherichicha coli. The overexpression of glycerol dehydrogenase in E. coli can be used as a way for endogenous mediator production. Many derivates of glycerol dehydrogenase are small, water-soluble redoxmolecules, which have mediator properties. In enzymology, a glycerol dehydrogenase (EC 1.1.1.6) is an enzyme belonging to the family of oxireductases that catalyzes the chemical reaction: Glycerol + NAD+ <--> Glycerone + NADH + H+ with NADH as the main endogenous mediator.
Theory
- Theory for GldA glycerol dehydrogenase.
Genetic Approach
- The GldA gene from Escherichia coli was cloned and heterologously expressed in E. coli KRX under the control of different promoters (Table 1).
Results
- Upon the expression of the glycerol dehydrogenase, the endogenous mediator production of Escherichia coli was measured. SDS-PAGE combined with MALDI-TOF MS/MS and different NADH-assays characterize GldA BioBrick <bbpart>BBa_K1172201</bbpart>.
SDS-PAGE and MALDI-TOF
- SDS-PAGE shows protein Glycerol dehydrogenase at expected size of 40 kDa. In contrast to Escherichia coli KRX Wildtyp, weak Anderson promoter (<bbpart>BBa_K1172205</bbpart>) shows only a slightly stronger band, whereas T7 (<bbpart>BBa_K1172203</bbpart>) and Lac (<bbpart>BBa_K1172204</bbpart>) promotor show a strong band, which is equated with a strong expression and overproduction of GldA.

Figure 2: SDS-PAGE with Prestained Protein Ladder from Thermo Scientific as marker. Comparison of protein expression between Escherichia coli KRX wild type and Escherichia coli KRX with <bbpart>BBa_K1172205</bbpart>, <bbpart>BBa_K1172204</bbpart> and <bbpart>BBa_K1172203</bbpart> after total cell disruption. The right-hand gel was loaded with a higher protein concentration. SDS-PAGE shows protein Glycerol dehydrogenase at expected size of 40 kDa. Enhanced overproduction with increasing promotor strength.
- Furthermore we were able to identify the overexpressed glycerol dehydrogenase (Figure. 2) with MALDI-TOF MS/MS.
- Tryptic digest of the gel lane for analysis with MALDI-TOF could examine the glycerol dehydrogenase with a Mascot Score of 266 against Escherichia coli database.
NADH-Assays
- An overproduction of the glycerol dehydrogenase results of course in an overproduction of products from glycerol dehydrogenase. Glycerol dehydrogenase produces several mediators because of its broad substrate specificity. GldA is involved in several metabolism pathways, for example in the glycerol metabolism, which converts glycerol and NAD+ to Glycerone, NADH and H+. NADH is a small, water-soluble redoxmolecule, which seems to be a great mediator for Microbial Fuel Cells. An overexpression of glycerol dehydrogenase leads to an overexpression of NADH and therefore to an endogenous mediator production. Furthermore GldA seems to be very interesting in combination with our preferred MFC carbon source glycerol. Thus, it is essential to get reliable data on NADH overproduction to see how many electron shuttles can be available and how efficient electron shuttle-mediated electron transfer (EET) will be.
- We could observe an enhanced NADH production for Escherichia coli KRX with pSB1C3 and <bbpart>BBa_K1172203</bbpart>, <bbpart>BBa_K1172204</bbpart> and <bbpart>BBa_K1172205</bbpart> with increasing promotor strength and in comparison with Escherichia coli KRX Wildtyp. (Table 2 and Figure 3)
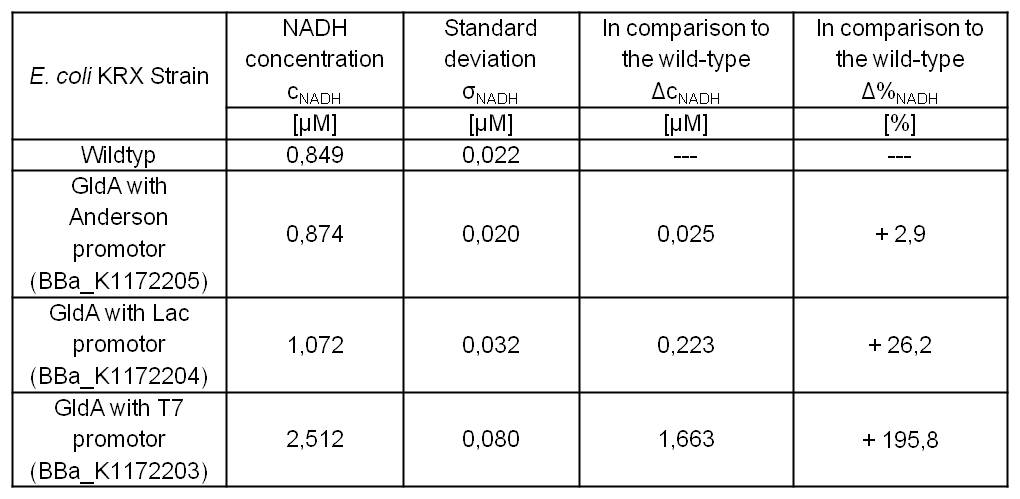
Table 2: Results of the NADH-Assay. Comparison between Escherichia coli KRX Wildtyp and Escherichia coli KRX with <bbpart>BBa_K1172205</bbpart>, <bbpart>BBa_K1172204</bbpart> and <bbpart>BBa_K1172203</bbpart>. NADH concentration with standard deviation and concentration in comparison to the wild-type is shown.
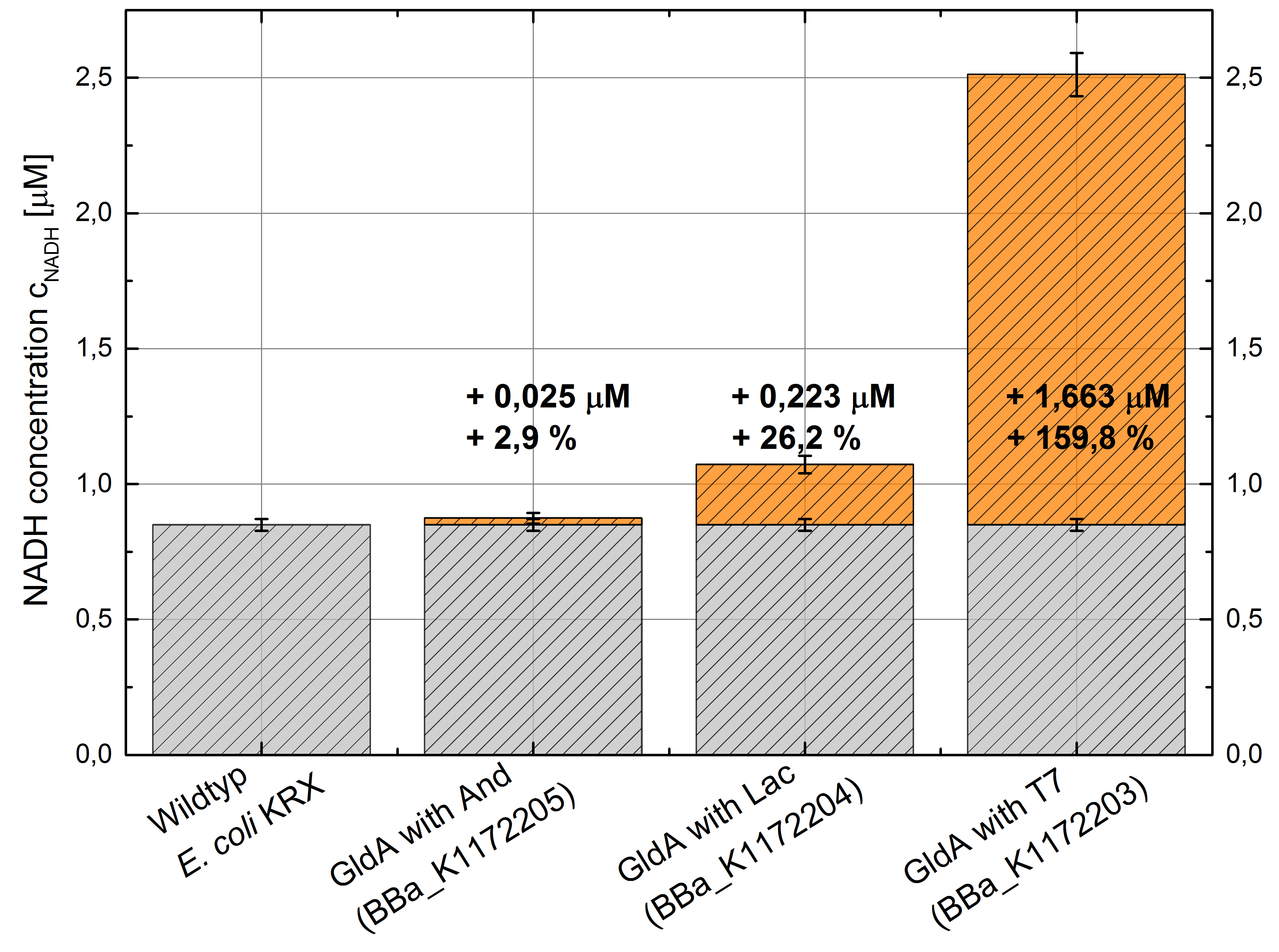
Figure 3: Column Chart with results of the NADH-Assay. Comparison between Escherichia coli KRX Wildtyp and Escherichia coli KRX with <bbpart>BBa_K1172205</bbpart>, <bbpart>BBa_K1172204</bbpart> and <bbpart>BBa_K1172203</bbpart>. NADH concentration with standard deviation and concentration in comparison to the wild-type is shown.
- Compared with Escherichia coli KRX Wildtyp, Escherichia coli with GldA and T7 promotor (<bbpart>BBa_K1172203</bbpart>) shows 1,7 μM NADH overproduction and GldA with Lac promotor (<bbpart>BBa_K1172204</bbpart>) shows 0,2 μM NADH overproduction. The GldA expression by the Anderson promotor (<bbpart>BBa_K1172205</bbpart>) is too weak for an efficient NADH overproduction.
References
- xxx
 "
"


