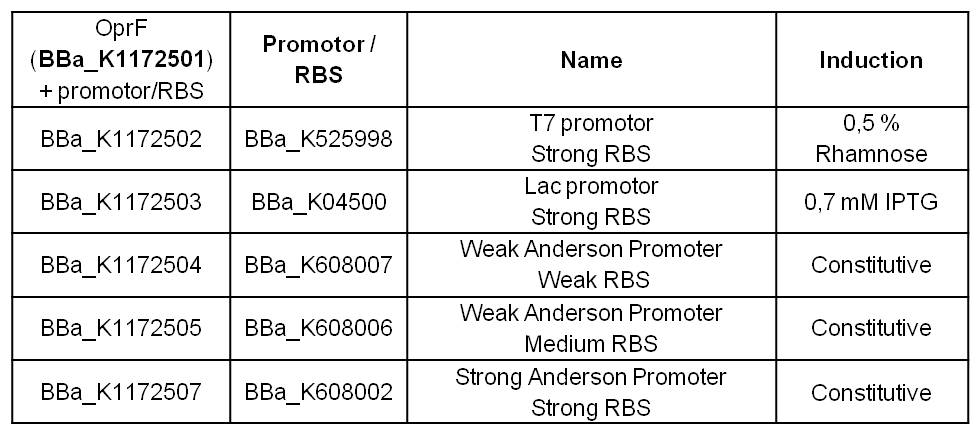
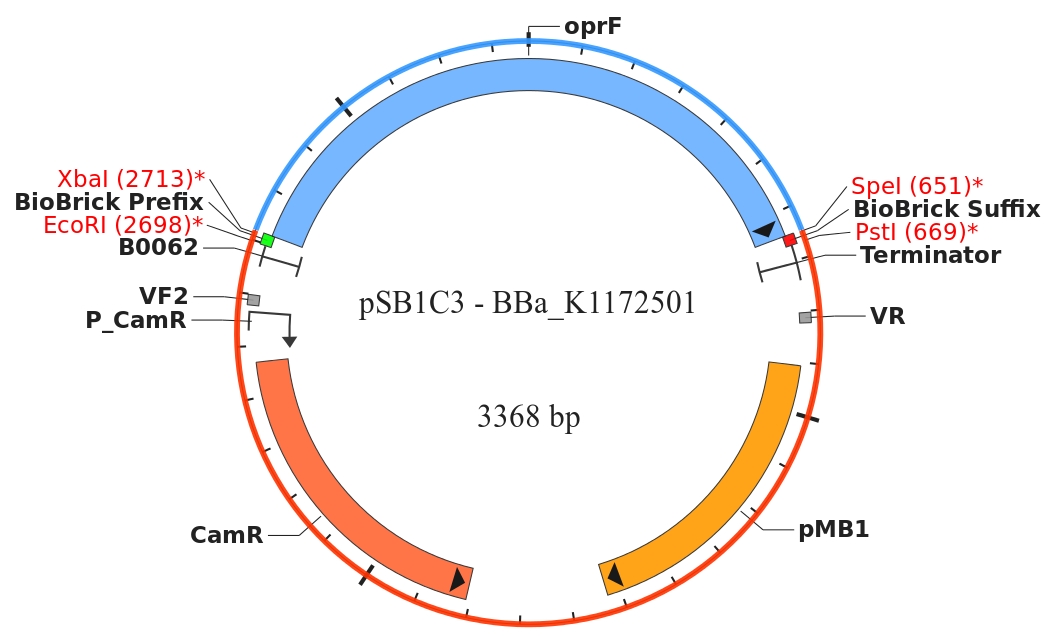
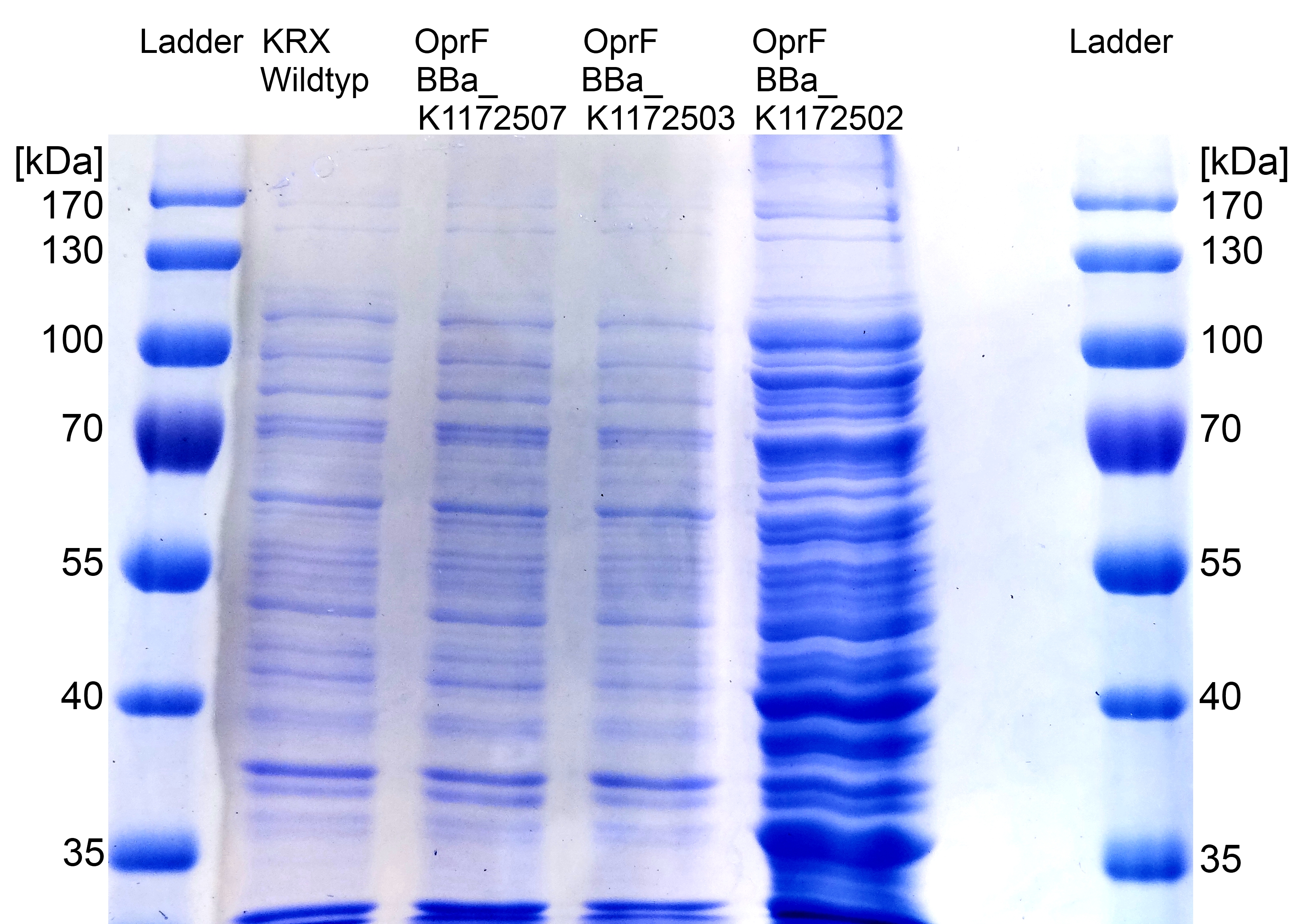
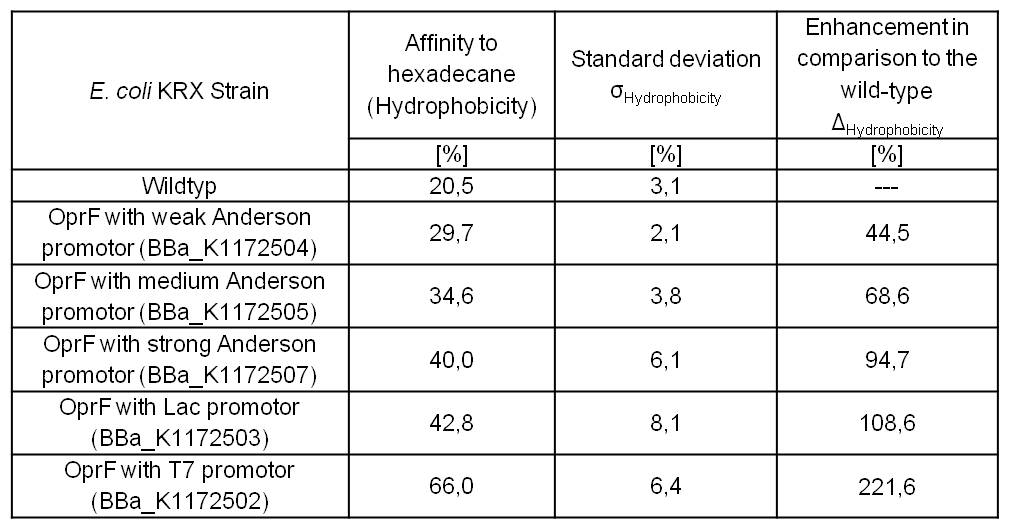
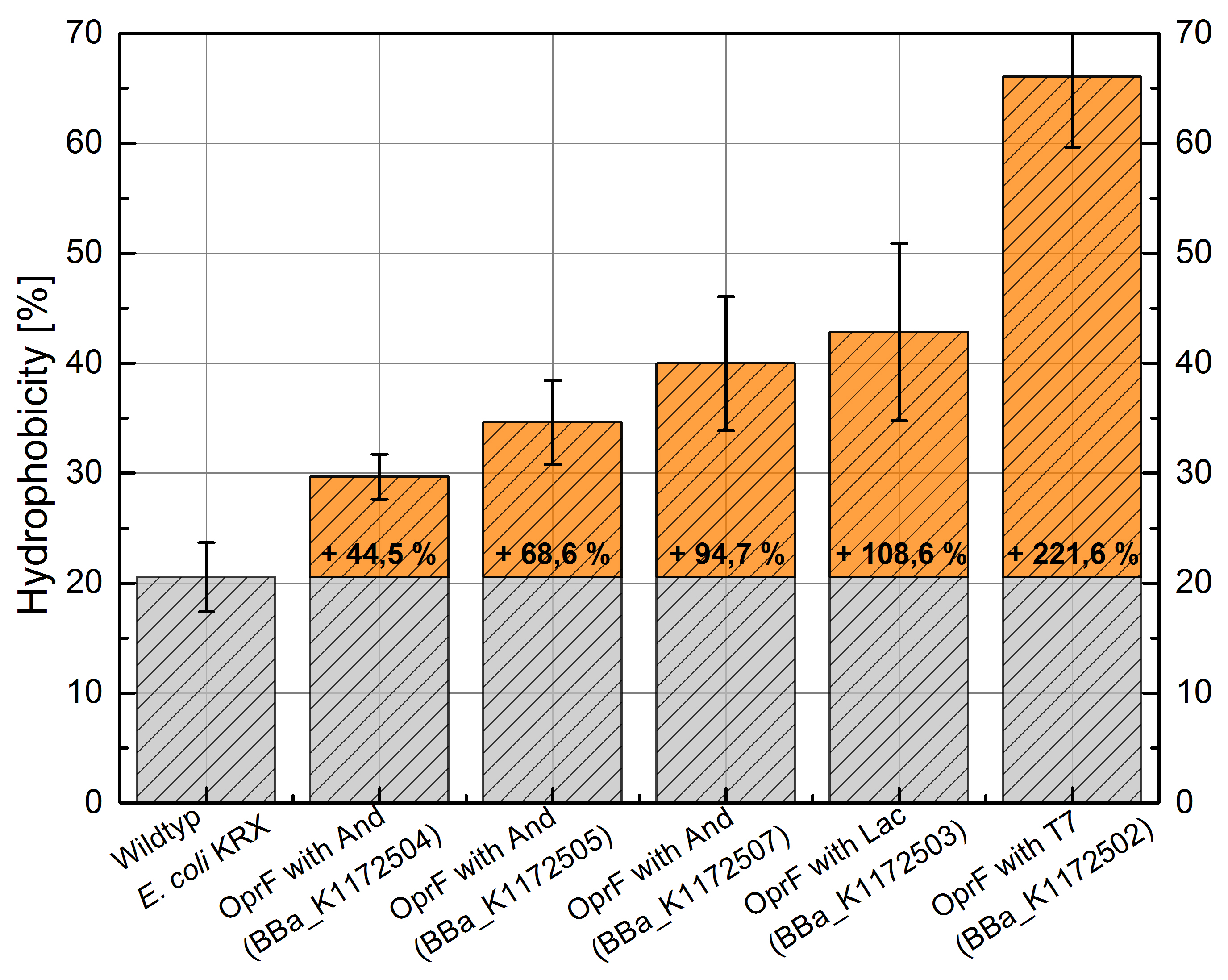
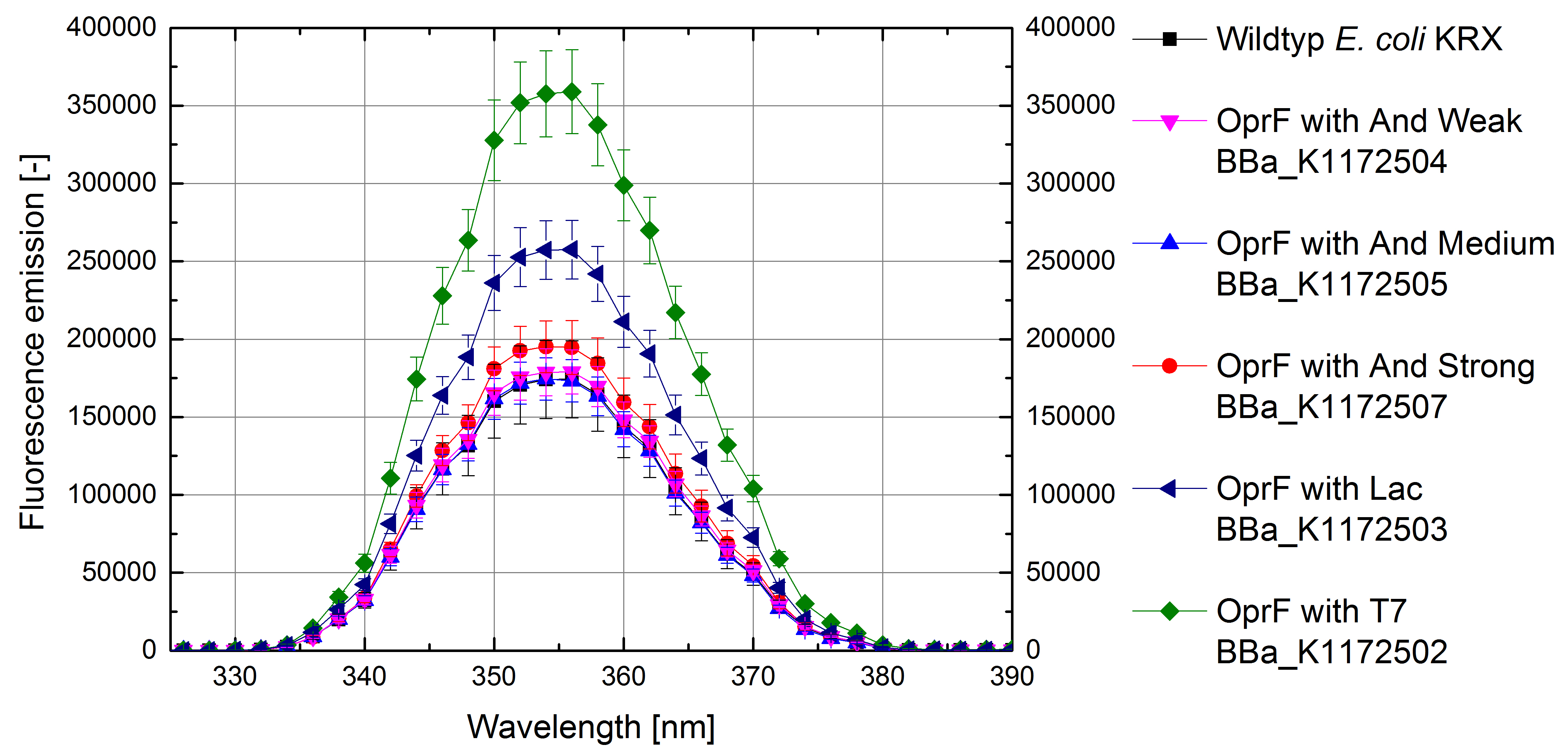
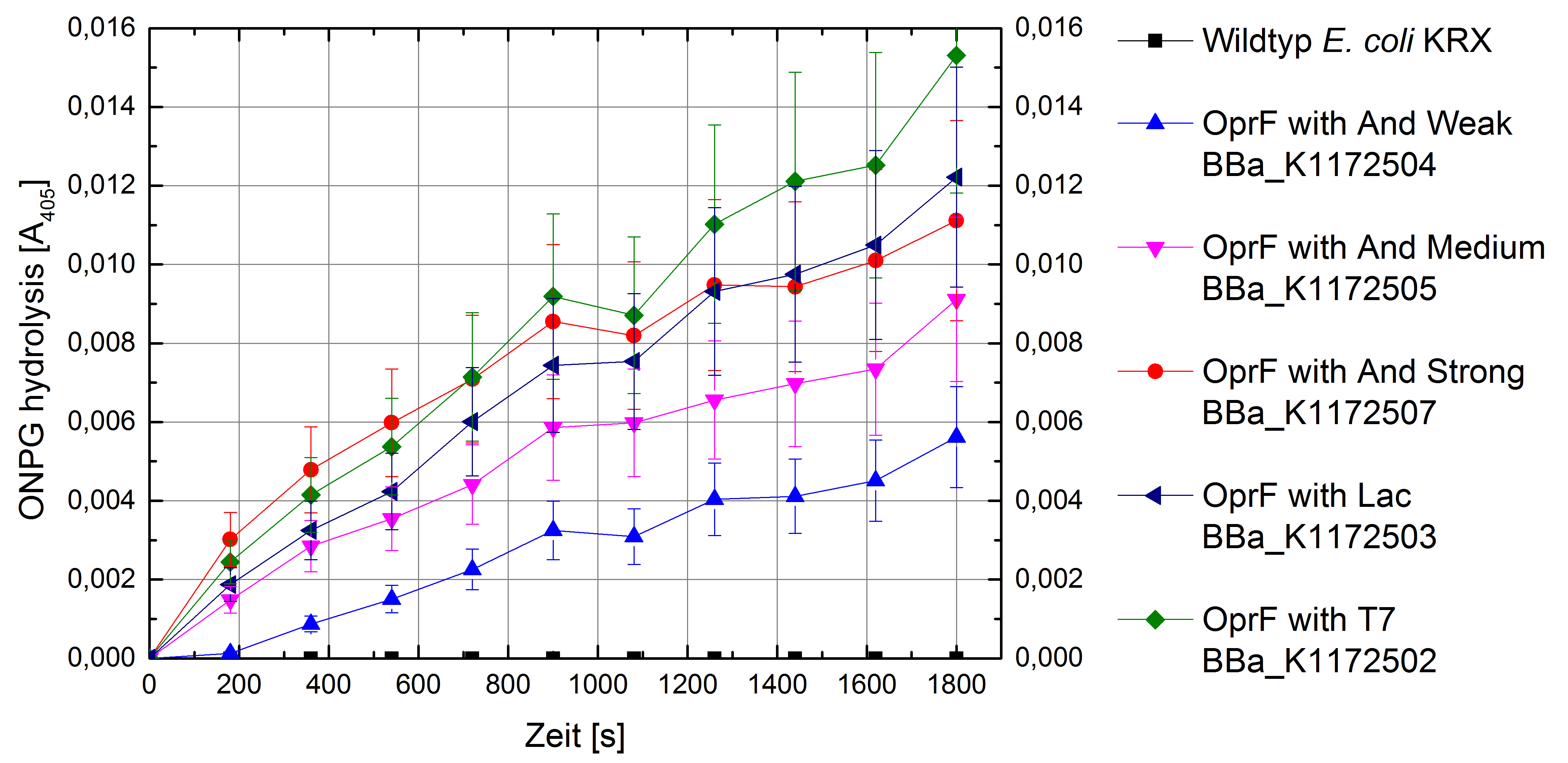
By heterologous expression of the pore-forming transmembrane-protein (or porine) OprF from Pseudomona fluorescens a further optimization of the E. coli membrane will be achieved. E. coli possesses several own porines, for example OmpF and OmpC. But these naturally occurring porines are only permeable for molecules smaller than 600 Da, which decreases the range of usable mediators significantly. Opposed to that, porine OprF from P. fluorescence forms one of the biggest known pores in the outer bacterial membrane with a permeability of up to 3000 Da, thus improving the electron shuttle mediated extracellular electron transfer (ETT) and enabling the usage of alternative mediators such as riboflavin (vitamine B2).
 "
"