Team:Heidelberg/Materials
From 2013.igem.org
| Line 13: | Line 13: | ||
} | } | ||
</style> | </style> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div class="container"> | <div class="container"> | ||
<div class="row" style="float:left"> | <div class="row" style="float:left"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div class="col-sm-12 jumbotron"> | <div class="col-sm-12 jumbotron"> | ||
<ol style="font-size:10px"> | <ol style="font-size:10px"> | ||
| - | + | </html>{{:Team:Heidelberg/Templates/Materials_list}}<html> | |
| - | + | ||
| - | + | ||
| - | + | ||
</ol> | </ol> | ||
Revision as of 21:52, 4 October 2013


- 0.5 g Yeast Extract (Difco-BectonDickinson)
- 1.0 g Cas Amino Acids (Difco)
- 2.0 g Pyruvic Acid
- 2.0 g L-Glutamine
- 0.3 g KH2PO4
- 0.3 g MgSO4
- 2.0 g MOPS
- 4.0 g Chelex 100-resin (Sigma)
- PH adjusted to 7.2–7.3 with 5 M KOH
- Fill up to 900 ml before adding pyruvic acid and L-glutamine
- Adjust pH
- Fill up to 1L
- Treat with Chelex for 1h
- Remove Chelex by filtration
- Add 200 ml of sterile M9 salt solution to 750 ml sterile, distilled H2O (45-50°C)
- Add sterile 20 ml 20% Glucose-solution, 2 ml 1 M MgSO4 and (optionally) 1 M CaCal
- 5.0 g Peptone
- 3.0 g Meat extract
- 1000.0 ml Distilled water
Kits
| Kit | Supplier | Catalog Number |
|---|---|---|
| MinElute® PCR Purification Kit (250) | QIAGEN | 28006 |
| Plasmid Plus Maxi Kit (25) | QIAGEN | 12963 |
| Plasmid Plus Midi Kit (25) | QIAGEN | 12943 |
| QIAEX II® Gel Extraction Kit (500) | QIAGEN | 20051 |
| QIAGEN® Plasmid Plus Midi Kit (100) | QIAGEN | 12945 |
| QIAquick® Gel Extraction Kit (250) | QIAGEN | 28706 |
| QIAquick® Nucleotid Removal Kit (250) | QIAGEN | 28306 |
| QIAquick® PCR Purification Kit (250) | QIAGEN | 28106 |
| QIAprep® Spin Miniprep Kit (250) | QIAGEN | 27106 |
| QIAprep® Spin Miniprep Columns | QIAGEN | 27115 |
Marker
| Marker | Supplier | Catalog Number |
|---|---|---|
| Quick-Load® 2-Log DNA Ladder (0.1-10.0 kb) | New England BioLabs | N3200S |
| Quick-Load® 1 kb DNA Ladder | New England BioLabs | N0468S |
| 50 bp DNA Ladder | New England BioLabs | N3236S |
| Gel loading solution | Sigma-Aldrich Chemie GmbH | G2526-5ML |
Enzymes
| Enzyme | Supplier | Catalog Number |
|---|---|---|
| DreamTaq Green PCR Master Mix (2X) | Thermo Fisher Scientific Biosciences GmbH | K1081 |
| DreamTaq PCR MM | Fermentas Life Sciences | K1071 |
| Gibson Assembly® Master Mix | New England Biolabs | E2611 S |
| Lysozyme from Chicken Egg White | Sigma-Aldrich Chemie GmbH | L4919-500MG |
| Phusion® Flash High-Fidelity PCR Master Mix | Biozym Scientific GmbH | F-548L |
| Phusion® High-Fidelity PCR Master Mix | New England Biolabs | M0531 L |
| T4 DNA Ligase | New England Biolabs GmbH | M0202 S |
| 2x PCR Master mix Solution (iTaq) | HISS DIAGNOSTICS GmbH | 25028 |
| Pronase from Streptomyces griseus | Sigma-Aldrich Chemie GmbH | P6911-100MG |
Restriction Enzymes
| Enzyme | Supplier | Catalog Number |
|---|---|---|
| BamI | New England Biolabs | R3136 S |
| BgIII | New England Biolabs | R0144 S |
| DpnI | New England Biolabs | R0176 S |
| EcoRI | New England BioLabs | R0101S |
| EcoRI-HF | New England Biolabs | R3101 |
| HindIII-HF | New England Biolabs | R3104 S |
| KpnI-HF | New England Biolabs | R3142 S |
| MfeI-HF | New England Biolabs | R3589 S |
| NheI-HF | New England BioLabs | R3131 S |
| NotI-HF | New England BioLabs | R3189 S |
| PacI | New England Biolabs | R0547 S |
| PstI-HF | New England Biolabs | R3140 S |
| PvuI-HF | New England BioLabs | R3150S |
| SalI-HF | New England Biolabs | R3138 S |
| SpeI-HF | New England BioLabs | R3133 S |
| XbaI | New England BioLabs | R0145 S |
Bacterial Strains
| Strain | Supplier | Catalog Number |
|---|---|---|
| Delftia acidovorans DSM-39 | DSMZ | DSM 50251 |
| Delftia acidovorans SPH-1 | DSMZ | DSM 14801 |
| E. coli DH10ß | New England Biolabs | C3019 |
| E. coli Top10 | invitrogen | C404010 |
| Photorhabdus laumondii luminescens | DSMZ | DSM 15139 |
| Streptomyces lavendulae lavendulae | DSMZ | DSM 40708 |
| Streptomyces mobaraensis | DSMZ | DSM 40903 |
Antibiotics and Media Supplements
| Antibiotic | Supplier | Catalog Number | Concentration stock solution | Dilution | Solvent |
|---|---|---|---|---|---|
| Ampicillin Anhydrous Crystalline | Sigma-Aldrich Chemie GmbH | A9393-5G | 100 mg/ml | 1:1000 | H2O |
| Ampicillin Sodium Crystalline | Sigma-Aldrich Chemie GmbH | A9518-5G | 100 mg/ml | 1:1000 | H2O |
| Chloramphenicol Crystalline | Sigma-Aldrich Chemie GmbH | C0378-5G | 30 mg/ml | 1:3000 | Ethanol |
| Kanamycinsulfat Mixture of Componenta | Sigma-Aldrich Chemie GmbH | 60615-5G | 50 mg/ml | 1:1000 | H2O |
| Tetracycline | Sigma-Aldrich Chemie GmbH | T7660 | 10 mg/ml | 1:1000 | Ethanol |
| Propionic Acid Sodium Insect Cell*Culture | Sigma-Aldrich Chemie GmbH | P5436-100G | 100mM | 10mM | H2O |
| Bacitracin | Sigma-Aldrich Chemie GmbH | B0125-50KU | - | - |
Media
| Medium | Supplier | Catalog Number |
|---|---|---|
| SOC Outgrowth Medium | New England Biolabs GmbH | B9020 S |
| LB BROTH BASE | Th. Geyer GmbH & Co KG | SA/L3022/001000 |
| LB Broth Powder | Sigma-Aldrich Chemie GmbH | L3022-1KG |
| M9 Minimal Salts | SERVA | 48505.01 |
Buffers
| Buffer | Supplier | Catalog Number |
|---|---|---|
| NEBuffer Pack #4 (green) | New England Biolabs GmbH | B7004 S |
| NEBuffer Pack #1 (yellow) | New England Biolabs GmbH | B7001 S |
| NEBuffer Pack for T4 DNA Ligase | New England Biolabs GmbH | B0202 S |
| NEBuffer Pack #2 (blue) | New England Biolabs GmbH | B7002 S |
| NEBuffer Pack #3 (red) | New England Biolabs GmbH | B7003 S |
| TAE - Buffer (50X) for Molecular Biology | VWR International GmbH | A4686.1000 |
| Gel Loading Buffer | Sigma-Aldrich | G2526-5ML |
| Tris Acetate-EDTA Buffer | Sigma-Aldrich | T9650-1L |
Other Chemicals
| Chemical | Supplier | Catalog Number |
|---|---|---|
| Isopropyl B-D-Thiogalactopyranoside 1 piece | Sigma-Aldrich Chemie GmbH | I5502-1G |
| Dimethyl Sulfoxide PCR Reagent | Sigma-Aldrich Chemie GmbH | D9170-1VL |
| Glycerol Sigma Grade | Sigma-Aldrich Chemie GmbH | G9012-100ML |
| 5-Bromo-4-Chloro-3-Indolyl B-D-*Galactop | Sigma-Aldrich Chemie GmbH | B4252-100MG |
| Bacteriological Agar | Sigma-Aldrich Chemie GmbH | A5306-250G |
| L-Plus-Arabinose Crystalline | Sigma-Aldrich Chemie GmbH | A3256-25G |
| Calciumchlorid Dihydrat | Th. Geyer GmbH & Co KG | SA/00223506/000500 |
| Malt Extract from Starch Digestion | Sigma-Aldrich Chemie GmbH | M0383-100G |
| D(+)-Saccharose, ACS, for Micro-Biology | Sigma-Aldrich Chemie GmbH | 84100-1KG |
| Dimethyl Sulfoxide Plant Cell Culture*TE | D4540-100ML | |
| Sodium Hydroxide Anhydrous Pellets | Th. Geyer GmbH & Co.KG | SA/S5881/000500 |
| TRIZMA(R) Hydrochloride PH 3.5-5.0 | Sigma-Aldrich Chemie GmbH | T6666-50G |
| L-Glutamine 200 MM Sterile | Sigma-Aldrich Chemie GmbH | G7513-20ML |
| Ethanol 96% Denatured | Carl Roth GmbH & Co.KG | T171.3 |
| Propanol-2 | Sigma-Aldrich Chemie GmbH | 59309-1L |
| Natriumdodecylsulfat,SDS,99%, Ultra Pure | 13904 | |
| Gold(III)-Chloride | Carl Roth GmbH & Co.KG | 5624.1 |
| Pro-Leu | Sigma-Aldrich Chemie GmbH | P1130-1G |
| Nitric Acid 65% p.a. Iso | Carl Roth GmbH & Co.KG | X943.1 |
| Mops, Sodium | Sigma-Aldrich Chemie GmbH | M9024-25G |
| L-Glutamine | Sigma-Aldrich Chemie GmbH | G7513-100ML |
| Chelating Resin | Sigma-Aldrich Chemie GmbH | C7901-50G |
| Potassium Hydroxide in Platellets | 6751.3 | |
| Hydrochloric Acid 37% | 4625.1 | |
| Pyruvic Acid | Sigma-Aldrich Chemie GmbH | 107360-25G |
| Fmoc-Orn(BOC)-OH 96.0 % | 47560-5G-F | |
| Glycerol >99.5% | Sigma-Aldrich Chemie GmbH | G9012-1L |
| Water Molecular Biology Reagent | Sigma-Aldrich Chemie GmbH | W4502-1L |
| Acetonitrile | Sigma-Aldrich Chemie GmbH | 34967-1L |
| Ascorbic Acid 99% | Sigma-Aldrich Chemie GmbH | A92902-100G |
Electrophoresis
| Reagent | Supplier | Catalog Number | Concentration | Solvent |
|---|---|---|---|---|
| Agarose Molecular Biology Reagent | Th. Geyer GmbH & Co KG | SA/A9539/000050 | 0.5% | H2O |
| Agarose for Routine Use | Sigma-Aldrich Chemie GmbH | A9539-100G | - | - |
Miscellaneous Primers
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| VF2 | - | for screening in standard BB backbone- binds on the Backbone before Insert | TGCCACCTGACGTCTAAGAA |
| VR | - | for screening in standard BB backbone- binds on Backbone behind Insert | ATTACCGCCTTTGAGTGAGC |
Primers and Oligos
Delftibactin
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| DN01:delH_f1_PacI_fw | - | fw Primer for DelH-fragment1 with RBS and PacI-restriction site | TTTT TTAATTAA TCACACAGGAAAGTACTAG ATGGACCGTGGCCGCCTGC GCCAAATCG |
| DN02:delH_f1_SalI_rev | - | - | TTTT GTCGACCAACACCTGTGCCTGC |
| DN03:delH_f2_SalI_fw | - | - | TTTT GTCGACTGGATGGAGCCTGGTGAAAG |
| DN04:delH_f2_KpnI_rev | - | - | TTTT GGTACC TCAGTCCAGCGCGTACTCCAG |
| DN05:AraCbb_KpnI_fw | - | amplifying the Backbone for DelH (pSB6A1-AraC-lacZ) | TTTT GGTACC AAAGAGGAGAAATACTAGATGACCATG |
| DN06:AraCbb_PacI_rev2 | 15-05-2013 | amplifying the Backbone for DelH (pSB6A1-AraC-lacZ) | TTTT TTAATTAA GCTAGCCCAAAAAAACGGTATG |
| DN07:Screen_delH_rev | 15-05-2013 | for screening if DelH is present - binds on the very beginning of DelH | CTTTCCTCGAACACCGTGCGCAG |
| DN08:DelH_EcoRI_rev | - | rev_Primer for DelH Fragment f1a | CTCGTCGCCATGGACCAGGCAG |
| DN09:DelH_f1_fw_long | 2013-06-11 | for amplifying DelH-1a from the genome: doesn't work | ATGGACCGTGGCCGCCTGCGCCAAATCG |
| DN10:DelH_f1_fw_short | 2013-06-11 | for amplifying DelH-1a from the genome: doesn't work | ATGGACCGTGGCCGCCTGC |
| DN11:DelH_f1_fw_short2 | 2013-06-11 | for amplifying DelH-1a from the genome: works!!!! | GCCGCCTGCGCCAAATCG |
| DN12:DelH_f1_PacI_fw_short | 2013-06-11 | for amplifying DelH-1a from the genome: doesn't work | TTTTTTAATTAATCACACAGGAAAGTAC TAGATGGACCGTGGCCGCCTGC |
| DN13:Screen_DelH_fw | 15-05-2013 | PCR Screening for presence of DelH insert | GTAAACCCACTGGTGATACCATTC |
| FS_01: pSB4K5_DelA_rv | 20-13-06-28 | Amplification of pSB4K5 from the iGEM Distribution Gibson Primer with overhang to DelA introducing the RBS BBa_B0035 | TCGCGGCGATCCGGTACTGCGCCTCTGTT GAACATCTGATATTCTCCTCTTTAATCG ACAGATTGTGTGAAATTGTTATCCGCTCAC |
| FS_02: DelAG_1_fw | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | TTCAACAGAGGCGCAGTACCGGATC |
| FS_03: DelAG_1_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | GTCGGAGACGATGTGGTGCATCAC |
| FS_04: DelAG_2_fw | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | CTGCAGGCCAATGAGCACATCCTG |
| FS_05: DelAG_2_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | CACAGGTGGTAGATGGCGTC |
| FS_06: DelAG_3_fwG | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | ATTGCGAGGACTTGCTCGATG |
| FS_07: DelAG_3_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | TTTGCTGCAGCGCCAGCACATCGAG |
| FS_08: DelAG_4_fw | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | GTACGGCCTATCACATCAGCG |
| FS_09: DelAG_4_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | GAAGCTCAGCAGGTTGGGCGAGACG |
| FS_10: DelAG_5_fw | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | GAATTTTGTTCCACCACCTGCTG |
| FS_11: DelAG_5_rv | 2013-06-28 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer with overhang to DelOP | CTTGAGCAGGCGCAGTACCTCGGAGGG CGGTCGGCTGGCGTTTTCCATGATT CAGGTTTCCTGTGTGAAGCTCATCTCAGATA TCTCCCAGAGTTTCGAGAAAG |
| FS_11: DelAG_5_short_rv | 2013-05-07 | Amplification of DelAG from Delftia acidovorans genome Gibson Primer | TCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_12: DelOP_fw | 2013-06-28 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | GAATCATGGAAAACGCCAGCCGAC |
| FS_13: DelOP_rv | 2013-06-28 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer with overhang to DelL | CAATGTTGGAGGGGCCGAAGCCGATGCCGATC AGCGGGTGGGTTTGCATGGAAGGTC CTTTCATTGGGTCGATGCGTCCAGTGT CACACCGTGGTGTCTGCAGGCG |
| FS_13: DelOP_short_rv | 2013-05-07 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer with overhang to DelL | TCACACCGTGGTGTCTGCAGGCG |
| FS_14: DelL_fw | 2013-06-28 | Amplification of DelL from Delftia acidovorans genome Gibson Primer | CAAACCCACCCGCTGATCGGCATC |
| FS_15: DelL_mRFP_pSB4K5_rv | 2013-06-28 | Amplification of DelL Gibson Primer with overhang to BBa_J04450 | GAAACGCATGAACTCTTTGATAACGTCT TCGGAGGAAGCCAT CTAGTATTTCTCCT CTTTCTCTAGTATCAGTCCTGCAGCG CCAGCTGTTCTGTG |
| FS_15: DelL_mRFP_pSB4K5__short_rv | 2013-05-07 | Amplification of DelL Gibson Primer with overhang to BBa_J04450 | TCAGTCCTGCAGCGCCAGCTGTTCTGTG |
| FS_16: mRFP_pSB4K5_fw (Del) | 2013-06-28 | Amplification of pSB4K5 from iGEM Distribution Gibson Primer | GCTTCCTCCGAAGACGTTATC |
| FS_20: DelF_fw | 2013-07-13 | Amplification of DelF from Delftia acidovorans genome Gibson Primer | GACTTGCTCGATGCGGTGCAG |
| FS_21: DelF_fw | 2013-07-13 | Amplification of DelF from Delftia acidovorans genome Gibson Primer | GACGCCATCTACCACCTGTG |
| FS_22: DelOP_short_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome inlcuding the recently predicted endogenous Promotor for DelOP Gibson Primer | GATGACGCAGGGCGGCGGAATTTGTTCATC |
| FS_23: DelG_long_rv | 2013-07-13 | Amplification of DelG from Delftia acidovorans genome Gibson primer with overhang to DelOP element including the recently predicted endogenous Promotor | GATGAACAAATTCCGCCGCCCTGCGTCA TCTCAGATATCTCCCAGAGTTTCGAGAAAG |
| FS_24: DelAE_rv | 2013-07-13 | Amplification of DelAE from Delftia acidovorans genome Gibson Primer | CAGAAGAATTCCCAGAAGGAGATGTCGAAG |
| FS_25: DelEF_fw | 2013-07-13 | Amplification of DelEF from Delftia acidovorans genome Gibson Primer | ACACGGTGCTGCAGAAAACGCCCTTC |
| FS_26: DelFG_rv | 2013-07-13 | Amplification of DelFG from Delftia acidovorans genome Gibson Primer | GAATTCATCCACGATGATCTGCATG |
| FS_27: DelOP_rv | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | CTTTGGGCCGTGCCGGTTTTTGAGATAC |
| FS_28: DelOP_rv | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | GTTTTTGAGATACGCGCGTTGTCAC |
| FS_29: DelOP_rv | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | TTCCCCCTCTCTTTCTCGCTTC |
| FS_30: DelOP_rv | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | CCGCTTCCCCCTCTCTTTCTCGCTTC |
| FS_31: DelOP_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | GTTGGCGAGTTCAAGAAATG |
| FS_32: DelOP_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | TCCTTCAGGTGTGCGGCAGACAAG |
| FS_33: DelOP_fw | 2013-08-02 | Amplification of DelOP from Delftia acidovorans genome Gibson Primer | TTCTTCGTGATGACGCAGGGCGGCGGAATTTGTTC |
| FS_35: DelG_fw | 2013-08-05 | Amplification of DelG from Delftia acidovorans genome Gibson Primer | CATGCAGATCATCGTGGATGAATTC |
| FS_45: pSB4K5_fw | 2013-08-02 | Amplification of pSB4K5 from iGEM Distribution Gibson Primer without mRFP | CCAGGCATCAAATAAAACGAAAG |
| FS_46: DelL_rv | 2013-08-02 | Amplification of DelL from Delftia acidovorans genome Gibson Primer creating overlap to pSB4K5 without mRFP | TCAGTCCTGCAGCGCCAGCTGTTCTG TGCTTTCGTTTTATTTGATGCCT |
| FS_47_screening_BB_AF_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTGGCCGATTCATTAATGC |
| FS_48_screening_BB_AF_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TAACGGTATCGGTATCGCTTTG |
| FS_49_screening_AFI_AFII_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTTCTCTGGAAGATGGATAC |
| FS_50_screening_AFI_AFII_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTGACGAAAAAGCCGACCAC |
| FS_51_screening_AF_FG(21-26)_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TGGATATCGACTGGACTGCCTG |
| FS_52_screening_AF_FG(21-26)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | TGCACCACATCGACGAAACGG |
| FS_53_screening_FG(21-26)_G_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTACGGCCTATCACATCAGCG |
| FS_54_screening_FG(21-26)_G_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GAACCTGGGTGTTCACGAAAAAGCC |
| FS_55_screening_G_OP_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTATCTCTACATGCATCGCTAC |
| FS_56_screening_G_OP_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | AGGACATTTTCCGCACCCCG |
| FS_57_screening_G_OP_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GCTGGCGTTTTCCATAAG |
| FS_58_screening_OP_L_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GAACAACTTCCAGCACAGCCTGTTC |
| FS_59_screening_OP_L_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CGTTGAAGATTTCGTTGACG |
| FS_60_screening_L_BB_fw | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CATCTTCAAGGTGTTCTATGAAC |
| FS_61_screening_L_BB(with_mRFP)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | CAGTTTAACTTTGTAGATGAAC |
| FS_66: DelH_rv | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | TGGGCATTCACCGCATCGATC |
| FS_67: DelH_fw | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | CTTCACGTTGATTGCGCATG |
| FS_68: DelH_rv | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | CAGAAGAACTCCCAGACCGAC |
| FS_69: DelH_fw | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | GACACCGTTCAGCTTCGATG |
| FS_70: DelH_rv | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | GAAGCTGCTCCGCTGATAGAT |
| FS_71: DelH_fw | 2013-08-26 | Amplification of DelH from Delftia acidovorans genome Gibson Primer | ATGTGCTGTCGCTCAAGATG |
| FS_72_SR_02_fw | 2013-08-30 | Screening of pFHFSN | ATGTGCTGTCGCTCAAGATG |
| FS_73_SR_03_fw | 2013-08-30 | Screening of pFHFSN | GTGCTGTTTGGCCGTATG |
| FS_74_SR_04_fw | 2013-08-30 | Screening of pFHFSN | ATCAGGTGCTGAGCTACGAC |
| FS_75_SR_05_fw | 2013-08-30 | Screening of pFHFSN | CTGTTCATCAACACCTTGCC |
| FS_76_SR_06_rv | 2013-08-30 | Screening of pFHFSN | GAAGACAGTCATAAGTGCGGC |
| FS_77_rv | 2013-09-11 | Gibson-Primer rev, Amplficiation of the Backbone pSB6A1 with overlap to the RBS BBa_B0034 and the lacI-promotor, it creates an overlap to the beginning of DelH | GCGATTTGGCGCAGGCGGCCACGGTC CATCTAGTATTTCTCCTCTTTC |
| FS_78_rv | 2013-09-26 | Gibson-Primer rev, Amplficiation of the Backbone pSB6A1 introducing the RBS BBa_B0032 and the promotor BBa_J23114 and creating an overlap to the first fragment of DelH amplified with primer DN_11 | GATTTGGCGCAGGCGGCCACGGTCCA TCTAGTACTTTCCTGTGTGACTCTAG AGCTAGCATTGTACCTAGGACTGAGCTAG CCATAAACTCTAGAAGCGGCCGCGAATTC |
| FS_84_fw | 2013-09-26 | Gibson-Primer fw, Amplficiation of the first fragment of DelH introducing the RBS BBa_B0032 and creating an overlap to primer FS_85 thereby partially introducing the promotor BBa_J23114 | GCTCAGTCCTAGGTACAATGCTAGCT CTAGAGTCACACAGGAAAGTACTAGA TGGACCGTGGCCGCCTGCG |
| FS_85_rv | 2013-09-26 | Gibson-Primer rev, Amplficiation of the Backbone pSB6A1, partially introducing the promotor BBa_B0032 with overlap to primer FS_84 and therefore the promotor BBa_J23114, it creates an overlap to the beginning of DelH | GCGATTTGGCGCAGGCGGCCACGGTCC ATCTAGTATTTCTCCTCTTTC |
| FS_86_rv | 2013-09-27 | Gibson-Primer rev, Amplficiation of the Backbone pSB4K5 without any promotor introducing a KpnI cutting site for restriction cloning, creates an overlap to DelH and will be used for the ccdB strategy | GGCGATTTGGCGCAGGCGGCCACGG TCCATGTACTTCGAGTCACTAAGGGCTAAC |
| FS_87_fw | 2013-09-27 | Gibson-Primer fw, Amplficiation of the Backbone pSB6A1 introducing a BamHI cutting site for restriction cloning and creating an overlap to the last fragment of DelH | CGCTGGAGTACGCGCTGGACTGA GATCCCAGGCATCAAATAAAACG |
| FS_90_fw | 2013-09-27 | Gibson-Primer fw, Amplficiation of the ccdB cassette from the template pDonorPlasmid introducing a KpnI cutting site for restriction cloning, creates an overlap to the promotor BBa_J23114 and will be used for the ccdB strategy | CTCAGTCCTAGGTACAATGCTAGCTCTAGA GTCACACAGGAAAGCAGTACACTGGCT GTGTATAAGGGAG |
| FS_93_rv | 2013-09-27 | Gibson-Primer rev, Amplficiation of the ccdB cassette from the template pDonorPlasmid introducing a BamHI cutting site for restriction cloning, creates an overlap to the backbone pSB6A1 and will be used for the ccdB strategy | GTTCACCGACAAACAACAGATGA TCCGCGTGGATCCGGCTTAC |
| FS_94_fw | 2013-09-27 | Primer fw, Amplficiation of the backbone pSB6A1, will be used for the ccdB strategy | ATCTGTTGTTTGTCGGTGAACGC |
| HM01:DelH_EcoRI_fw | - | fw_Primer for DelH Fragment f1b | GCATTGGAGCCTCAATGGCAAGTC |
| HM02:DelH_Gib1.1_rev | 2013-07-09 | Gibson-Primer DelH | TGCTGCGCCTGCATACGGCCAAACA |
| HM03:DelH_Gib1.2_fw | 2013-07-09 | Gibson-Primer DelH | AGCGGCAGGGACGACGTGGT |
| HM04:DelH_Gib1.2_rev | 2013-07-09 | Gibson-Primer DelH | CATAGAGGTTGTAGAGA |
| HM05:DelH_Gib2.1_fw | 2013-07-09 | Gibson-Primer DelH | AGAACGCCGTCTTCAGGCTCCTG |
| HM06:DelH_Gib2.1_rev | 2013-07-09 | Gibson-Primer DelH | CAATGCTTTG CCGCTCGAA |
| HM07:DelH_Gib2.2_fw | 2013-07-09 | Gibson-Primer DelH | TCGCCACGGCAGCTGTTCGA |
| HM08:DelH_Gib2_end_rev | 2013-07-09 | Gibson-Primer DelH | TCAGTCCAGCGCGTACTCCAG |
| HM09:AraC_RBS_Delh_rev | 2013-07-09 | Gibson-Primer rev, introduces a new RBS and has the AraC-promotor and the beginning of DelH | TTGCAAAGCGCTCGGCGATTTGGCGCAGGCG GCCACGGTCCATTTAACTTTCTCCTC TTTAATACTTTGAGCTAGCCCAA AAAAACGGTATGGAGAAACAGTAGAGAGTT |
| HM10:RBS_lacZ | 2013-07-09 | Gibson-Primer fw for the pSB6A1 Backbone with the end of DelH, RBS(1) and the beginning of lacZ | TGGAGTACGCGCTGGACTGA TCTAGAG AAAGAGGAGAAA TACTAG ATGACCATGATTA |
| HM11:lacI_RBS(1)_DelH_rev | 2013-07-24 | Gibson-Primer rev, amplify the Backbone with overlap with the RBS and the lacI-promotor and it creates and overlap to the start of DelH | TCGGCGATTTGGCGCAGGCGGCCACGGTCC ATCTAGTATTTCTCCTCTTTCTCTAGTATGTGTG |
| HM12:DelH_RBS(1.2)_mRFP_fw | 2013-07-24 | Gibson-Primer fw for the pSB6A1 Backbone with the end of DelH, introducing a new RBS(new) and the beginning of mRFP | ATTGGCGCTGGAGTACGCGCTGGACTG ATCAAAGTATTAAAGAGGA GAAAGT TAAATGGCTTCCTCCGAAGACGTTATCAAAGAG |
| HM13:Screen_DelH_end_fw | 2013-08-16 | New screening primer for the end of DelH together with the VR2 primer from the registry | TTTCTGACGACCCTGCACCTGAAG |
| HM14:DelH_tetR_fw | 2013-08-16 | Gibson-Primer DelH-tetR: amplifies the tetracycline resistance from the pSB1T3 Backbone and creates an overlap to the end of DelH | ATTGGCGCTGGAGTACGCGCTGGACTGA ATGAAGTTTTAAATCAATCTAAAG |
| HM15:tetR_stop_BB_rev | 2013-08-16 | Gibson-Primer tetR-pSB6A1: amplifies the tetracycline resistance and creates an overlap with the Terminator of the Backbone pSB6A1 | CGACTGAGCCTTTCGTTTTATTTGATGCCTGGC CTCGTGATACGCCTATTTTTATAGG |
| HM16:tetR_pSB6A1_fw | 2013-08-16 | Gibson-Primer DelH, amplifies the Backbone pSB6A1 creating an overlap with the tetracycline resistance | AAAAATAGGCGTATCACGAG GCCAGGCA TCAAATAAAACGAAAGGCTCAG |
| HM17:DelH_Terminator_BB_fw | 2013-08-16 | Gibson-Primer fw for the pSB6A1 Backbone (binding the terminator) and creating an overlap with the end of DelH | ATTGGCGCTGGAGTACGCGCTGGACTGA AGGCATCAAATAAAACGAAAGGCTCAG |
| HM20:BB_HPLC_rev | 11-09-2013 | HPLC version of HM11 Gibson-Primer rev, amplify the Backbone with overlap with the RBS and the lacI-promotor and it creates and overlap to the start of DelH | GATTTGGCGCAGGCGGCCAC GGTCCATCTAGTATTTCTCCTCTTTC |
| HM21:fw_lacI_BbsI_Xba | 2013-09-15 | Forward primer for cutting out mutated fragment for mutagenesis | TTTTGAAGACAA CTAGGCAATACGCAA |
| HM22:rev_RBS | 2013-09-15 | Reverse Primer in RBS for mutagenesis | TTTTGAAGACAA CTCTTTCTCTAGTATGTGTGAAATTG |
| HM23:fw_RBS | 2013-09-15 | Forward Primer in RBS for mutagenesis | TTTTGAAGACAA AGAGGAGAAATACTAGATGGACCGTGGC |
| HM24:rev_BbsI_MfeI | 2013-09-15 | Reverse primer for cutting out mutated fragment for mutagenesis | TTTTGAAGACAA AATTGGACAGCGCGGCATGCCGGTTG |
| IK01:pLF03_integr_argK_fw | 2013-06-12 | For verification of correct genomic integration of pLF03 into E.coli BAP1 via colony-PCR. Primer against E.coli BAP1 genome. | GCTGATGGAAGTGGCTGATCTGATC |
| IK02:pLF03_integr_pET21c_rev | 2013-06-12 | For verification of correct genomic integration of pLF03 into E.coli BAP1 via colony-PCR. Primer against pLF03 backbone (pET-21c). | TCCGCTCACAATTCCCCTATAGTG |
| IK03:pLF03_integr_argK_rev | 2013-06-18 | For verification of correct genomic integration of pLF03 into E.coli BAP1 via colony-PCR. Primer against E.coli BAP1 genome for positive control (to be used with IK01 or IK04). | GATAAATTCACTGAGCTGCCGCAG |
| IK04:pLF03_integr_argK_fw | 2013-06-18 | For verification of correct genomic integration of pLF03 into E.coli BAP1 via colony-PCR. Primer against E.coli BAP1 genome. Alternative forward primer in case IK01 does not work. | GCGGGAATTAATGCTGTTATGCGAAG |
| IK05:pLF03_integr_ygfG_fw | 2013-06-18 | For verification of correct genomic integration of pLF03 into E.coli BAP1 via colony-PCR. Primer against E.coli BAP1 genome, namely ygfG, which should be replaced by the construct. | GCGGTCATTGAGTTTAACTATGGCC |
| IK06:pLF03_integr_ygfG_rev | 2013-06-18 | For verification of correct genomic integration of pLF03 into E.coli BAP1 via colony-PCR. Primer against E.coli BAP1 genome, namely ygfG, which should be replaced by the construct. | AACATGGTTGAGGATGCCGACAGC |
| IK07:ygfG21C1 | 2013-06-25 | For insertion of methylmalonyl-CoA synthesis pathway into E. coli. Extended ygfG21C1 with higher melting temperature. | TCACCGCGCACCGGCCTGCGGCAGCTCAGTGAATTTATCC AGATCTCGATCCCGCGAAATTAATAC |
| IK08:ygfG21C2 | 2013-06-25 | For insertion of methylmalonyl-CoA synthesis pathway into E. coli. Extended ygfG21C2 with higher melting temperature. | GTTATGCTGGATAATTTCTGCCGCTTCATTGGCGGTCATC CAAAAAACCCCTCAAGACCCGTTTAG |
| IK24:pccB_fw | 2013-07-16 | Colony-PCR of methylmalonyl-CoA synthesis pathway | CGTATCGAGGAAGCGACGCAC |
| IK25:pccB_rev | 2013-07-16 | Colony-PCR of methylmalonyl-CoA synthesis pathway | GGTGATGAACATGTGGCTGGTCTG |
| IK26:BBa_I746200_fw | 2013-07-19 | Forward primer for permeability device | ATGAACAAGAAGATTCATTCCCTGGCCTTG |
| IK27:BBa_I746200-BBa_B0029_rev | 2013-07-19 | Reverse primer for permeability device with partial Gibson overhang for BBa_B0029 RBS | GAATACCAGT TCAGAAGTGGGTGTTTACGCTCATATAC |
| IK28:pccB-BBa_B0029-BBa_I746200_fw | 2013-07-19 | Forward primer for pccB with BBa_B0029 RBS and Gibson overhang for permeability device | ATATGAGCGTAAACACCCACTTCTGAACTGGTATTCACAC AGGAAACCTACTAG ATGTCCGAGCCGGAAGAGC |
| IK29:accA2-BBa_B0030_rev | 2013-07-19 | Reverse primer for accA2 with partial Gibson overhang for BBa_B0030 RBS | TAATGAAGTTG CTAGTGATTCTCGCAGATGGC |
| IK30:sfp-BBa_B0030-accA2_fw | 2013-07-19 | Forward primer for sfp with BBa_B0030 RBS and Gibson overhang for accA2 | TCCGGCGCCGCCATCTGCGAGAAT CACTAGCAACTTCATTAAAGAGGAG AAATACTAG ATGAAGATTTACGGAATTTATATGGAC |
| IK31:sfp_rev | 2013-07-19 | Reverse primer for sfp | TTATAAAAGCTCTTCGTACGAGAC |
| IK32:BBa_J04450-sfp_fw | 2013-07-19 | Forward primer for mRFP-containing backbones with Gibson overhang for sfp | ATCACAATGGTCTCGTACGAAGAGCTTTTATAA TACTAGAGCCAGGCATCAAATAAAACG |
| IK33:BBa_J04450-BBa_I746200_rev | 2013-07-19 | Reverse primer for mRFP-containing backbones with Gibson overhang for permeability device | ACAAGGCCAGGGAATGAATCTTCTTGTTCAT CTAGTATTTCTCCTCTTTCTCTAGTATG |
| IK34:pccB_fw | 2013-07-19 | Forward primer for pccB | ATGTCCGAGCCGGAAGAGC |
| IK35:BBa_J04450-pccB_rev | 2013-07-19 | Reverse primer for mRFP-containing backbones with Gibson overhang for pccB | ATGTCGGGCTGCTGCTCTTCCGGCTCGGACAT CTAGTATTTCTCCTCTTTCTCTAG |
| IK36:pLF03_seq_rev | 2013-07-26 | reverse primer for pLF03 for sequencing | CCGGTATCAACAGGGACACCAG |
| IK37:pLF03-catR_seq_rev | 2013-07-26 | reverse primer for pLF03 sequencing | CATTGAGCAACTGACTGAAATGCCTC |
| IK38:pIK1_fw | 2013-08-30 | Forward mutagenic primer for pIK1 | ATTCCCTGGCCTTGT T GGTCAATCTGGGGATTTATG |
| IK39:pIK_rev | 2013-08-30 | Reverse mutagenic primer for pIK1 | CCCAGATTGACC A ACAAGGCCAGGGAATGAATCTTC |
| IK40:pIK2-BBa_J23114- BBa_B0030_rev | 2013-09-06 | reverse primer for pIK2 with Gibson overhang for BBa_J23114-BBa_B0030 | TTTAATCTCTAGAGCTAGCATTGTACCTAGGACTGAGCTAGCCATAAA CTCTAGTAGAGAGCGTTCAC |
| IK41:BBa_I746200-BBa_B0030 -BBa_J23114_fw | 2013-09-06 | forward primer for BBa_I746200 with Gibson overhang for BBa_B0030-BBa_J23114 | ATGCTAGCTCTAGAGATTAAAGAGGAGAAATACTAG ATGAACAAGAAGATTCATTCCCTG |
| IK42:BBa_I746200_rev | 2013-09-06 | reverse primer for BBa_I746200 | TCAGAAGTGGGTGTTTACGCTC |
| IK43:pIK2-BBa_I746200_fw | 2013-09-06 | forward primer for pIK2 with Gibson overhang for BBa_I746200 | TGAGCGTAAACACCCACTTCTGA TACTAGAGTCACACTGGCTC |
| NK_01_FS_62_screening_L_ BB(without_mRFP)_rv | 2013-08-02 | Primer for screening/sequencing of pFSN construct | GTTCACCGACAAACAACAGATAAAACG |
| ygfG21C1 | 2013-06-03 | For insertion of methylmalonyl-CoA synthesis pathway into E. coli. Primer from <bib id="pmid17959404"/>. | TCACCGCGCACCGGCCTGCGGCAGCTCAGTGAATTTATCC AGATCTCGATCCC |
| ygfG21C2 | 2013-06-03 | For insertion of methylmalonyl-CoA synthesis pathway into E. coli. Primer from <bib id="pmid17959404"/>. | GTTATGCTGGATAATTTCTGCCGCTTCATTGGCGGTCATCC CAAAAAACCCCTCAAG |
Module Shuffling
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| AT01:RFC10prefix_TycA_fw | 2013-08-12 | Fw primer for amplification of TycAdCom; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG ATG TTA GCA AAT CAa GCC AAT C |
| AT02:RFC10suffix_TycA_rv | 2013-08-12 | Rv primer for amplification of TycAdCom; introduction of RFC10 suffix | TTTT CTGCAG CGGCCGC T ACTAGT A aGT TCG tTC TAC TTC TTT TTT C |
| AT03:RFC10prefix-TycB1_fw | 2013-08-12 | Fw primer for amplification of TycB1dCom; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG ATG AGT GTA TTT AGC AAA GAA CAA G |
| AT04:RFC10suffix_TycB1_rv | 2013-08-12 | Rv primer for amplification of TycB1dCom; introduction of RFC10 suffix | TTTT CTGCAG CGGCCGC T ACTAGT A TTC CTC CCC aCC TTC |
| AT05:RFC10prefix-TycC5_fw | 2013-08-12 | Fw primer for amplification of TycC5; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG AG GCG CAT ATT GCa GAG AG |
| AT06:RFC10suffix_TycC5_rv | 2013-08-14 | Rv primer for amplification of TycC5; introduction of RFC10 suffix | TTTT CTGCAG CGGCCGC T ACTAGT A TTT GGC TGT CTC TTC GAT GAA C |
| AT07:RFC10prefix-TycC6_fw | 2013-08-12 | Fw primer for amplification of TycC6; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG AG GGG AAT GTC TTC TCG ATC |
| AT08:RFC10suffix_TycC6_rv | 2013-08-14 | Rv primer for amplification of TycC6; introduction of RFC10 suffix | TTTT CTGCAG CGGCCGC T ACTAGT A TTA TTT CAG GAT aAA CAG TTC TTG |
| AT09:R10_B1_fw_longer | 2013-08-18 | Fw primer (longer) for amplification of TycB1dCom; introduction of RFC10 prefix | TTTT GAA TTC GCG GCC GCT TCT AG ATG AGT GTA TTT AGC AAA GAA CAA GTT C |
| AT10:R10_B1_rv_longer | 2013-08-18 | Rv primer (longer for amplification of TycB1dCom; introduction of RFC10 suffix | TTTT CTG CAG CGG CCG CTA CTA GTA ATA CGC aCT TTC CTC CCC GCC |
| AT11:R10_C5_fw_rpos | 2013-08-18 | Fw primer (repositioned) for amplification of TycC6; introduction of RFC10 prefix | TTTT GAATTC GCGGCCGC T TCTAG AG GAGCAGTTCGAGACGATCCAGCC |
| AT101 | 2013-09-05 | forward screening primer TycAdCom | GGACATCATCGAACAGGC |
| AT102 | 2013-09-05 | reverse screening primer TycAdCom | GACATAGGCAAGATCCGTAG |
| AT103 | 2013-09-05 | forward screening primer TycAdCom | GCAAGGACAAGTAGATGGC |
| AT104 | 2013-09-05 | reverse screening primer TycAdCom | GTACAGCCATACAGTCGC |
| AT105 | 2013-09-05 | forward screening primer TycC5 | GTCACCATGCTGAGAACG |
| AT106 | 2013-09-05 | reverse screening primer TycC5 | CAGCAAAGCGGTCATAATC |
| AT107 | 2013-09-05 | forward screening primer TycC6 | AAGCTACGCTGTTGATTGC |
| AT108 | 2013-09-05 | reverse screening primer TycC6 | AGCACGTAACGATCCTC |
| IK09:TycA_A1_fw | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycA A domain | ATGTTAGCAAATCAAGCCAATCTC |
| IK10:TycA_A1_rev | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycA A domain | TTGGTTTGCTGTAAGATCAGGCTC |
| IK11:TycB_A1_fw | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycB A1 domain | AATTCGGGAGTCGAGCTTTGTCAG |
| IK12:TycC6_rev | 2013-07-19 | reverse primer for tyrocidine TycC6 module | TTATTTCAGGATGAACAGTTCTTGCAGG |
| IK13:IK13:TycB1-dCom-dC_fw | 2013-07-19 | forward primer for tyrocidine TycB1 module without Com and C domain | GATTGCGTGGCAAACAATTCGGGAGTC |
| IK14:TycB1-TycC6_rev | 2013-07-19 | reverse primer for tyrocidine TycB1 module with Gibson overhang for TycC6 | CAGGCTCGATCGAGAAGACATTCCC TTCCTCCCCGCCTTCCACATACGC |
| IK15:TycC6-TycB1_fw | 2013-07-19 | forward primer for tyrocidine TycC6 module with Gibson overhang for TycB1 | GCGTATGTGGAAGGAGGGGAGGAA GGGAATGTCTTCTCGATCGAGCCTG |
| IK16:TycA_fw | 2013-07-19 | forward primer for tyrocidine TycA module | ATGTTAGCAAATCAGGCCAATCTCATC |
| IK17:TycA-dCom-TycC5_rev | 2013-07-19 | reverse primer for tyrocidine TycA module without Com domain with Gibson overhang for TycC5 | GAATGCGCTCTCGGCAATATGGGC TGTTCGCTCTACTTCTTTTTTCTCGG |
| IK18:TycC5-TycA-dCom_fw | 2013-07-19 | forward primer for tyrocidine TycC5 module with Gibson overhang for TycAdCom | CCGAGAAAAAAGAAGTAGAGCGAACA GCCCATATTGCCGAGAGCGCATTC |
| IK19:TycB1-TycC5_rev | 2013-07-19 | reverse primer for tyrocidine TycB1 module with Gibson overhang for TycC5 | GAATGCGCTCTCGGCAATATGGGC TTCCTCCCCGCCTTCCACATACGC |
| IK20:TycC5-TycB1_fw | 2013-07-19 | forward primer for tyrocidine TycC5 module with Gibson overhang for TycB1 | GCGTATGTGGAAGGAGGGGAGGAA GCCCATATTGCCGAGAGCGCATTC |
| IK21:pSB4K5-TycB1dComdC_rev | 2013-07-19 | reverse primer for mRFP-carrying backbones with Gibson overhang for tyrocidine TycA module (nomenclature wrong) | GTCGATGAGATTGGCCTGATTTGCTAACAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| IK22::pSB4K5-TycC6_fw | 2013-07-19 | forward primer for mRFP-carrying backbones with Gibson overhang for tyrocidine TycC6 module | AACATCCTGCAAGAACTGTTCATCCTGAAA TAATAACGCTGATAGTGCTAGTGTAGATC |
| IK23:pSB4K5-TycA_rev | 2013-07-19 | reverse primer for mRFP-carrying backbones with Gibson overhang for tyrocidine TycB1 module without Com and C domains + start codon (nomenklature wrong) | GACTCCCGAATTGTTTGCCACGCAATCCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW01:TycB_A1_rev | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycB A1 domain | CTTGGCACTTCCTTCAGGCTTC |
| PW02:TycB_E1_fw | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycB E domain | CGAGAGAGCGAGCAAGGTG |
| PW03:TycB_E1_rev | 2013-07-08 | Colony-PCR of Brevibacillus parabrevis: TycB E domain | GTACTCGCCTTCTTCTTTTGC |
| PW04:pSB1C3-TycC5ΔC_rev | 2013-07-19 | Integration of Tetrapeptide NRPS from Brevibacillus parabrevis in backbone; Gibson primer for Tetrapeptide I & II | TGTTTTGGTTGCGAGGAAGCTGTGCAGCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW05:TycC5ΔC_fwd | 2013-07-19 | Amplification of TycC5-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | ATGCTGCACAGCTTCCTCGCAACCAAAACAGCC |
| PW06:TycC5ΔC-TycB1ΔCom_rev | 2013-07-19 | Amplification of TycC5-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | GTGAAACAGCATCCCCTCTTGCATCGG AGGCTCGATCGAGAAGACATTCCCTTTG |
| PW07:TycC5ΔC-TycB1ΔCom_fwd | 2013-07-19 | Amplification of TycB1+C(TycB2)-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | CAAAGGGAATGTCTTCTCGATCGAGCCT CCGATGCAAGAGGGGATGCTGTTTCAC |
| PW08:C(TycB2)-TycAΔCom_rev | 2013-07-19 | Amplification of TycB1+C(TycB2)-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | GTTGTCGATGAGATTGGCCTGATTTGCTAACAT GATTTGCGCCAGCTCCTGCTCCGTGTT |
| PW09:C(TycB2)-TycAΔCom_fwd | 2013-07-19 | Amplification of TycA-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I & II | AACACGGAGCAGGAGCTGGCGCAAATC ATGTTAGCAAATCAGGCCAATCTCATCGACAAC |
| PW10:TycAΔCom-TycC6_rev | 2013-07-19 | Amplification of TycAΔCom-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I | CTTTGGCTGTCTCTTCGATGAACGC TCGCTCTACTTCTTTTTTCTCGGTGCAATG |
| PW11:TycAΔCom-TycC6_fwd | 2013-07-19 | Amplification of TycC6-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide I | CATTGCACCGAGAAAAAAGAAGTAGAGCGA GCGTTCATCGAAGAGACAGCCAAAG |
| PW12:TycAΔE-TycC6_rev | 2013-07-19 | Amplification of TycAΔE-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide II; one mismatch C->t | CTTTTGCACAGGCTCGATCGAGAAGAC GCTtTTGACAAAAAGAGCAACCTG |
| PW13:TycAΔE-TycC6_fwd | 2013-07-19 | Amplification of TycC6-Module from Brevibacillus parabrevis; Gibson primer for Tetrapeptide II; one mismatch G->a | CAGGTTGCTCTTTTTGTCAAaAGC GTCTTCTCGATCGAGCCTGTGCAAAAG |
| PW14:C(TycC2)-indC_rev | 2013-08-16 | Amplification of C-domain from TycC2 from Brevibacillus parabrevis; Gibson overhang to IndC; for construct 1, 2 & 3 | ACATTGTGTAATATTATTTTCTAACAT CGTTTTGCTGCTGGCAGGCTG |
| PW15:C(TycC2)-indC_fwd | 2013-08-16 | Amplification of indC from Photorhabdus luminescens; Gibson overhang to C-domain from TycC2; for construct 1, 2 & 3 | CAGCCTGCCAGCAGCAAAACG ATGTTAGAAAATAATATTACACAATGT |
| PW16:indC_rev | 2013-08-16 | Amplification of indC from Photorhabdus luminescens; no Gibson overhang; for construct 1, 2 & 3 | TTAGATTATTTTCTCAATCTCAGCAACACCTTC |
| PW17:TycAdE-C(TycC2)_rev | 2013-08-16 | Amplification of TycAdE from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC2; for construct 1 | CGAAAGGAAGCGGGCCAGCTC AGCAACCTGCTCGATCGTCGGGTA |
| PW18:TycAdE-C(TycC2)_fwd | 2013-08-16 | Amplification of C-domain from TycC2-module from Brevibacillus parabrevis; Gibson overhang to TycAdE; for construct 1 | TACCCGACGATCGAGCAGGTTGCT GAGCTGGCCCGCTTCCTTTCG |
| PW19:TycC4dC_fwd | 2013-08-16 | Amplification of TycC4-module from Brevibacillus parabrevis without the C-domain; no Gibson overhang, ATG added; for construct 3 | ATGTATCCGCGCGATCTGACGATTC |
| PW20:TycC4dC-C(TycC2)_rev | 2013-08-16 | Amplification of TycC4-module from Brevibacillus parabrevis without the C-domain; Gibson overhang to C-domain form TycC2; for construct 3 | GGTGTACTCGGTTTTTTCCGA AATATGCGCAGCCAACTCATG |
| PW21:TycC4dC-C(TycC2)_fwd | 2013-08-16 | Amplification of C-domain from TycC2-module from Brevibacillus parabrevis; Gibson overhang to TycC4dC; for construct 3 | CATGAGTTGGCTGCGCATATT TCGGAAAAAACCGAGTACACC |
| PW22:pSB1C3-TycC4dC_rev | 2013-08-16 | Insertion of construct 3 into pSB1C3-backbone, amplification of pSB1C3; Gibson overhang to TycC4dC; for construct 3 | GAATCGTCAGATCGCGCGGATACAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW23:indC-pSB1C3_fwd | 2013-08-16 | Insertion of either fragments into pSB1C3-backbone, amplification of pSB1C3; Gibson overhang to indC from Photorhabdus luminescens; for constructs 1, 2 & 3 | GGTGTTGCTGAGATTGAGAAAATAATCTAA TAATAACGCTGATAGTGCTAGTGTAGATC |
| PW24:TycC1dC_fwd | 2013-08-16 | Amplification of the TycC1-module from Brevibacillus parabrevis without the C-domain; no Gibson overhang, ATG added; for construct 2 | ATGCAGACGAACAAACAACAGACG |
| PW25:pSB1C3-TycC1dC | 2013-08-16 | Insertion of construct 2 in pSB1C3-backbone, amplification of pSB1C3; Gibson overhang to TycC1-module without C-domain; for construct 2 | CGTCTGTTGTTTGTTCGTCTGCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW26:TycC5dC_fwd | 2013-09-04 | Amplification of TycC5-module without C-domain from Brevibacillus parabrevis; no Gibson overhang, ATG added; for constructs A & B | ATGCTGCACAGCTTCCTCGCAACC |
| PW27:TycC5dC-C(TycC4)_rev | 2013-09-04 | Amplification of TycC5-module without C-domain from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC4; for constructs A & B | CACATACGTCTCTTTTCCGCTCGT TTCGATGAACGCCGCCAGTTC |
| PW28:TycC5dC-C(TycC4)_fwd | 2013-09-04 | Amplification of C-domain from TycC4-module from Brevibacillus parabrevis; Gibson overhang to TycC5-module without C-domain; for constructs A & B | GAACTGGCGGCGTTCATCGAA ACGAGCGGAAAAGAGACGTATGTG |
| PW29:C(TycC4)-TycC4dC_rev | 2013-09-04 | Amplification of C-domain from TycC4-module from Brevibacillus parabrevis; Gibson overhang to TycC4-module without C-domain; for constructs A, B, C, E & G | GAATCGTCAGATCGCGCGGATA GGCAAACGTGTTGTTGAAATC |
| PW30:C(TycC4)-TycC4dC_fwd | 2013-09-04 | Amplification of TycC4-module without C-domain from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC4-module; for constructs A, B, C, E & G | GATTTCAACAACACGTTTGCC TATCCGCGCGATCTGACGATTC |
| PW31:TycC4-C(TycC4)_rev | 2013-09-04 | Amplification of TycC4-module from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC4-module; for constructs B & C | CACATACGTCTCTTTTCCGCTCGT GGCAATATGCGCAGCCAACTCATG |
| PW32:TycC4-C(TycC4)_fwd | 2013-09-04 | Amplification of C-domain from TycC4-module from Brevibacillus parabrevis; Gibson overhang to TycC4-module; for constructs B & C | CATGAGTTGGCTGCGCATATTGCC ACGAGCGGAAAAGAGACGTATGTG |
| PW33:TycC6dTE-C(TycC2)_rev | 2013-09-04 | Amplification of TycC6-module without the TE-domain from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC2-module; for constructs D & F | GGTGTACTCGGTTTTTTCCGA CGTGATGAAATCGGCCACCTTTTC |
| PW34:TycC6dTE-C(TycC2)_fwd | 2013-09-04 | Amplification of C-domain from TycC2-module from Brevibacillus parabrevis; Gibson overhang to TycC6-module without TE-domain; for constructs D & F | GAAAAGGTGGCCGATTTCATCACG TCGGAAAAAACCGAGTACACC |
| PW35:TycC6dTE-C(TycC4)_rev | 2013-09-04 | Amplification of TycC6-module without the TE-domain from Brevibacillus parabrevis; Gibson overhang to C-domain from TycC4-module; for constructs E & G | CACATACGTCTCTTTTCCGCTCGT CGTGATGAAATCGGCCACCTTTTC |
| PW36:TycC6dTE-C(TycC4)_fwd | 2013-09-04 | Amplification of C-domain from TycC4-module from Brevibacillus parabrevis; Gibson overhang to TycC6-module without TE-domain; for constructs E & G | GAAAAGGTGGCCGATTTCATCACG ACGAGCGGAAAAGAGACGTATGTG |
| PW37:pSB1C3-TycC5dC_rev | 2013-09-04 | Insertion of constructs A & B in pSB1C3-backbone, amplification of pSB1C3; Gibson overhang to TycC5-module without C-domain; for constructs A & B | GGTTGCGAGGAAGCTGTGCAGCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
Linker Variation
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| AR01 | 2013-09-17 | sTycC4dC_fw | ATG AAGCACTTGCTGCCGCTCGTC |
| AR02 | 2013-09-17 | c(TycC2)s_rev | TTGTGTAATATTATTTTCTAACAT CGCTACCTGTTGCAACCGCTC |
| AR03 | 2013-09-17 | s-Ind_fw | GAGCGGTTGCAACAGGTAGCG ATGTTAGAAAATAATATTACAATGT |
| AR04 | 2013-09-17 | c(TycC2)l_rev_l | TTGTGTAATATTATTTTCTAACAT AAGCGACTGCTGGTTCGCG |
| AR05 | 2013-09-17 | l-Ind_fw | AACGCGAACCAGCAGTCGCTT ATGTTAGAAAATAATATTACACAATGT |
| AR06 | 2013-09-17 | lTycC4dC_fw | ATG CGCAACTATCCGGTCGAGACG |
| AR07 | 2013-09-17 | TycC4dCom_short-BB_rev | GACGAGCGGCAGCAAGTGCTTCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| AR08 | 2013-09-17 | TycC4dCom_long-BB_rev | CGTCTCGACCGGATAGTTGCGCAT CTAGTATTTCTCCTCTTTCTCTAGTATGTG |
| PW19:TycC4dC_fwd | 2013-08-16 | Amplification of TycC4-module from Brevibacillus parabrevis without the C-domain; no Gibson overhang, ATG added; for construct 3 | ATGTATCCGCGCGATCTGACGATTC |
| PW20:TycC4dC-C(TycC2)_rev | 2013-08-16 | Amplification of TycC4-module from Brevibacillus parabrevis without the C-domain; Gibson overhang to C-domain form TycC2; for construct 3 | GGTGTACTCGGTTTTTTCCGA AATATGCGCAGCCAACTCATG |
| PW21:TycC4dC-C(TycC2)_fwd | 2013-08-16 | Amplification of C-domain from TycC2-module from Brevibacillus parabrevis; Gibson overhang to TycC4dC; for construct 3 | CATGAGTTGGCTGCGCATATT TCGGAAAAAACCGAGTACACC |
| PW14:C(TycC2)-indC_rev | 2013-08-16 | Amplification of C-domain from TycC2 from Brevibacillus parabrevis; Gibson overhang to IndC; for construct 1, 2 & 3 | ACATTGTGTAATATTATTTTCTAACAT CGTTTTGCTGCTGGCAGGCTG |
Domain Shuffling and PPTases
| Identifier | Order date | Note | Sequence |
|---|---|---|---|
| KH1_ccdb_fw | 2013-07-23 | amplifing ccdb with RBS from pDONR | TGGTGCCATTACATACAGATACT GAGAACAGGGGCTGGTGAAATGC |
| KH2_ccdb_rv | 2013-07-23 | amplifing ccdb with RBS from pDONR | AGAGTCTGTCTGTTCAATCCACTT TTATATTCCCCAGAACATCAGGTTAATGGCG |
| KH3_indC_backbone-withoutT_fw | 2013-07-23 | linearization of vector with partial indigoidine synthase | AAGTGGATTGAACAGACAGACTCTAAAAC |
| KH4_indC_backbone-withoutT_rv | 2013-07-23 | linearization of vector with partial indigoidine synthase | AGTATCTGTATGTAATGGCACCAATAGACGC |
| KH5_indC_T_fw | 2013-07-23 | introducing T domain of indC | TGGTGCCATTACATACAGATACT GAAATAAGGCTTGGAAAAATTTGGATGGAAGT |
| KH6_indC_T_rv | 2013-07-23 | introducing T domain of indC | AGAGTCTGTCTGTTCAATCCACTT AGCCAATTCTGCTATATTAGGAGATTGA |
| KH7_bpsA_T_fw | 2013-07-23 | introducing T domain of bpsA | TGGTGCCATTACATACAGATACT GAGAAGGAAATCGCAGCCGTGTGGG |
| KH8_bpsA_T_rv | 2013-07-23 | introducing T domain of bpsA | AGAGTCTGTCTGTTCAATCCACTT GGCCAGCTTTTCAATTGTTGGTGACTCC |
| KH9_ccdB-Big_fw | 2013-08-06 | ccdB_Big amplified from pDONOR (Dominik) for indC(ccdB) construct | TGCCATTACATACAGATACT ACTGGCTGTGTATAAGGGAGCCTGAC |
| KH10_ccdB-Big_rv | 2013-08-06 | ccdB_Big amplified from pDONOR (Dominik) for indC(ccdB) construct | AGAGTCTGTCTGTTCAATCCACTT CGCGTGGATCCGGCTTAC |
| KH11_plu2642-T_fw | 2013-08-14 | T domain exchange plu2642 | TGGTGCCATTACATACAGATACT GAAACACAGATCGTAAAGATATGG |
| KH12_plu2642-T_rv | 2013-08-14 | T domain exchange plu2642 | AGAGTCTGTCTGTTCAATCCACTT TGCCAATTGTTTAACCGTTG |
| KH13_BBa-entD_fw | 2013-08-09 | submitting endD to registry | GCAT GAATTCGCGGCCGCTTCTAG ATGAAAACTACGCATACCTCCCTC |
| KH14_BBa-entD_rv | 2013-08-09 | submitting endD to registry | GCAT CTGCAGCGGCCGCTACTAGTA TCATTAATCGTGTTGGCACAGCG |
| KH15_BBa-sfp_Bsub_fw | 2013-08-09 | submitting sfp_Bsub to registry | GCAT GAATTCGCGGCCGCTTCTAG ATGAAGATTTACGGAATTTATATGGAC |
| KH16_BBa-sfp_Bsub_rv | 2013-08-09 | submitting sfp_Bsub to registry | GCAT CTGCAGCGGCCGCTACTAGTA TCATTATAAAAGCTCTTCGTACGAGAC |
| KH17_BBa-svp_Svert_fw | 2013-08-09 | submitting svp_Svert to registry | GCAT GAATTCGCGGCCGCTTCTAG A TGATCGCCGCCCTCCTGCCCTCC |
| KH18_BBa-svp_Svert_rv | 2013-08-09 | submitting svp_Svert to registry | GCAT CTGCAGCGGCCGCTACTAGTA AGCTAGCTCATTACGGGAC |
| KH19_BBa-delC_fw | 2013-08-09 | submitting delC to registry | GCAT GAATTCGCGGCCGCTTCTAG ATGCAGCTCGTGTCCGTGCG |
| KH20_BBa-delC_rv | 2013-08-09 | submitting delC to registry | GCAT CTGCAGCGGCCGCTACTAGTA TTA TCATGTCGATTCCTTGGTGC |
| KH21_BBa-indC_fw | 2013-08-09 | submitting indC to registry | GCAT GAATTCGCGGCCGCTTCTAG ATGTTAGAAAATAATATTACACAATG |
| KH22_BBa-indC_rv | 2013-08-09 | submitting indC to registry | GCAT CTGCAGCGGCCGCTACTAGTA TTAGATTATTTTCTCAATCTCAG |
| NI01:bpsA_AOxA_PstI_fw | 2013-06-04 | for amplifying bpsA (AOxA domain) | GAGGAGAAATACTAGATGACACTGCAGGAAACAAGCGTGC |
| NI02:bpsA_AOxA_rv | 2013-06-04 | for amplifying bpsA (AOxA domain) | GAAGGGCCGTTCCACCAGCTCAGCGTTGACCTGGTCAGAAGCG |
| NI03:bpsA_T_fw | 2013-06-04 | for amplifying bpsA (T domain) | GACCAGGTCAACGCTGAGCTGGTGGAACGGCCCTTCGTCG |
| NI04:bpsA_T_rv | 2013-06-04 | for amplifying bpsA (T domain) | GACGAAGCGACTAGACTCCTGAGCCACTTCTCTCTCCAGCCG |
| NI05:bpsA_TE_fw | 2013-06-04 | for amplifying bpsA (TE domain) with RBS 1 | GAGAGAGAAGTGGCTCAGGAGTCTAGTCGCTTCGTCCGA |
| NI06:bpsA_TE_Stop_XbaI_rbs_rv | 2013-06-04 | for amplifying bpsA (TE domain) with RBS 1 | TGGCAGCAGAGCAGCGATCATCTAGTATTT CTCCTCTTTCCTCTAGATCATCATTCCC CCAGCAGGTATCTAAT |
| NI07:svp_fw | 2013-06-04 | for amplifying svp | GAGGAGAAATACTAGATGATCGCTGCTCTGCTGCCAAGTTGG |
| NI08:svp_rv | 2013-06-04 | for amplifying svp | GCACTATCAGCGTTATTATGGCACGGCAGTCCTATCGTCG |
| NI09:pSB1C3_fw | 2013-06-04 | for linearizing, amplifying pSB1C3 | GATAGGACTGCCGTGCCATAATAA CGCTGATAGTGCTAGTGTAGATCGC |
| NI10:pSB1C3_PstI_rv | 2013-06-04 | for linearizing, amplifying pSB1C3 | GCTTGTTTCCTGCAGTGTCATCTAG TATTTCTCCTCTTTCTCTAGTATGTG |
| NI11:bpsA_rvN | 2013-07-15 | for amplifying bpsA (TE domain) with RBS 2 | TGGCAGCAGAGCAGCGATCATTATTTAGGT TTCCTGTGTGAATCATCATTCCCCCAGCAGGTATCTAAT |
| NI12:svp_fwN | 2013-07-15 | for amplifying svp with RBS 2 | CAGGAAACCTAAATAATGATC GCTGCTCTGCTGCCAAGTTGG |
| NI13_ccdb_fw | 2013-07-23 | amplifing ccdb with RBS from pDONR | CGTCGCACCTAGGACCGAAACA GAGAACAGGGGCTGGTGAAATGC |
| NI14_ccdb_rv | 2013-07-23 | amplifing ccdb with RBS from pDONR | GCCACTTCTCTCTCCAGCCGTCG TTATATTCCCCAGAACATCAGGTTAATGGCG |
| NI15_bpsA_backbone-withoutT_fw | 2013-07-23 | linearization of vector with partial indigoidine synthase | CGACGGCTGGAGAGAGAAGTGGC |
| NI16_bpsA_backbone-withoutT_rv | 2013-07-23 | linearization of vector with partial indigoidine synthase | TC TGTTTCGGTCCTAGGTGCGACG |
| NI17_indC_T_fw | 2013-07-23 | introducing T domain of indC | CGTCGCACCTAGGACCGAAACA GAAATAAGGCTTGGAAAAATTTGGATGGAAGT |
| NI18_indC_T_rv | 2013-07-23 | introducing T domain of indC | GCCACTTCTCTCTCCAGCCGTCG AGCCAATTCTGCTATATTAGGAGATTGA |
| NI19_bpsA_T_fw | 2013-07-23 | introducing T domain of bpsA | CGTCGCACCTAGGACCGAAACA GAGAAGGAAATCGCAGCCGTGTGGG |
| NI20_bpsA_T_rv | 2013-07-23 | introducing T domain of bpsA | GCCACTTCTCTCTCCAGCCGTCG GGCCAGCTTTTCAATTGTTGGTGACTCC |
| NI19:ngrA_fw | GAATTCGCGGCCGCTTCTAG ATGGAACAGACAGTTATACACACC | ||
| NI20:ngrA_rv | CTGCAGCGGCCGCTACTAGTA TTATTGAATAATTAGCGTTAATATTTCCTG | ||
| RB05:RBS1-BpsA_fw | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | AAAGAGGAGAAA TACTAG ATGACACTTCAGGAAACCAGCG |
| RB06:BpsA-RBS2_rv | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | CTTTCCTGTGTGA AAGCTT TCA TCACTCCCCGAGCAGATATCG |
| RB07:RBS2-svp_fw | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | CTTTCACACAGGAAAG TAAATA ATGGCTGCTCTTCTTCCTAGTTGGGC |
| RB08:svp-pSB1C3_rv | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | CTATCAGCGTTATTA AAGCTT TCATCA TGGCACGGCAGTCCTATCG |
| RB09:pSB1C3_fw | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | AAGCTT TAATAACGCTGATAGTGCTAGTGTAGATCGC |
| RB10:pSB1C3-RBS1_rv | 2013-07-01 | Gibson assembly of pSB1C3 bpsA svp | CTAGTA TTTCTCCTCTTT CTCTAGTATGTGTG |
| RB11:BpsA_Taka_fw | 2013-07-08 | colony PCR bpsA Takahashi | CAT ATGACTCTTCAGGAGACCAGCGTGCTC |
| RB12:BpsA_Taka_rv | 2013-07-08 | colony PCR bpsA Takahashi | AAG CTTCTCGCCGAGCAGGTAGCGGATGTG |
| RB13:svp_Taka_fw | 2013-07-08 | colony PCR svp Takahashi | CAT ATGATCGCCGCCCTCCTGCCCTCCTG |
| RB14:svp_Taka_rv | 2013-07-08 | colony PCR svp Takahashi | CTCGAGCGGGACGGCGGTCCGGTCGTCCGC |
| RB15:svp_Sanch_fw | 2013-07-08 | PCR svp Sanchez | TATA ATGCATGCTCGCCGCCCTCCCC |
| RB16:svp_Sanch_rv | 2013-07-08 | PCR svp Sanchez | TTAAGATCTCGGGACGGCGGTCCGGTC |
| RB17:sfp_fw | 2013-07-08 | sfp extraction from E.coli BAP1 | ATGAAGATTTACGGAATTTATATGG |
| RB18:sfp_rv | 2013-07-08 | sfp extraction from E.coli BAP1 | TTATAAAAGCTCTTCGTACGAGACC |
| RB19:entD_fw | 2013-07-08 | entD extraction from E.coli Lambalot | TAAATA ATGGTCGATATGAAAACTACGC |
| RB20:entD_rv | 2013-07-08 | entD extraction from E.coli Lambalot | AAGCTT ATTAATCGTGTTGGCACAGCG |
| RB21:pSB1C3_fw | 2013-07-15 | indigoidine exchangeable construct | TAATGA GCTAGC TAATAACGCTGATAGTGCTAGTG |
| RB22:pSB1C3_rv | 2013-07-15 | indigoidine exchangeable construct | CAT GGTACC TTTCTCCTCTTT CTCTAGTATGTGTG |
| RB23:bpsA-fus_fw | 2013-07-15 | indigoidine exchangeable construct | CACACATACTAGAG AAAGAGGAGAAA GGTACC ATGACTAGTACACTGCAGG |
| RB24:bpsA-fus_rv | 2013-07-15 | indigoidine exchangeable construct | CAT GGATCC GGTTTCCTGTGTGAA TCATTA TTCCCCCAGCAGGTATCTAATATG |
| RB25:svp-fus_fw | 2013-07-15 | indigoidine exchangeable construct | CACAGGAAACC GGATCC ATGACTAGTATCGCTGCTCTGCTG |
| RB26:svp-fus_rv | 2013-07-15 | indigoidine exchangeable construct | CAGCGTTATTA GCTAGC TCATTA TGGCACGGCAGTCCTATC |
| RB27:indC-Plum_fw | 2013-07-15 | indigoidine exchangeable construct | CATACTAGAG AAAGAGGAGAAA GGTACC ATGTTAGAAAATAATATTACACAATG |
| RB28:indC-Plum_rv | 2013-07-15 | indigoidine exchangeable construct | CAT GGATCC GGTTTCCTGTGTGAA TTA TTAGATTATTTTCTCAATCTCAG |
| RB29:svp-Svert_fw | 2013-07-15 | indigoidine exchangeable construct | CACAGGAAACC GGATCC A TGATCGCCGCCCTC |
| RB30:svp-Svert_rv | 2013-07-15 | indigoidine exchangeable construct | CAGCGTTATTA GCTAGC TCA TTACGGGACGGCGGTC |
| RB31:Sc-indC-Schr_fw | 2013-07-15 | indigoidine exchangeable construct | GAG AAAGAGGAGAAA GGTACC ATGAGCGTAGAGACCATC |
| RB32:Sc-indC-Schr_rv | 2013-07-15 | indigoidine exchangeable construct | CAT AGATCT GGTTTCCTGTGTGAA TTA TCAGTAGTTGGGCGTCTTG |
| RB33:entD-MG1655_fw | 2013-07-15 | indigoidine exchangeable construct | CACAGGAAACC GGATCC ATGAAAACTACGCATACCTC |
| RB34:entD-MG1655_rv | 2013-07-15 | indigoidine exchangeable construct | CAGCGTTATTA GCTAGC TCA TTAATCGTGTTGGCACAGC |
| RB35:sfp-Naka_fw | 2013-07-15 | indigoidine exchangeable construct | CACAGGAAACC GGATCC ATGAAGATTTACGGAATTTATATGGAC |
| RB36:sfp-Naka_rv | 2013-07-15 | indigoidine exchangeable construct | CAGCGTTATTA GCTAGC TCA TTATAAAAGCTCTTCGTACGAGAC |
| RB37:Plum-extr_fw | 2013-07-15 | genomic extraction | ATGTTAGAAAATAATATTACACAATG |
| RB38:Plum-extr_rv | 2013-07-15 | genomic extraction | TTAGATTATTTTCTCAATCTCAG |
| RB39:Svert-extr_fw | 2013-07-15 | genomic extraction | GTGATCGCCGCCCTC |
| RB40:Svert-extr_rv | 2013-07-15 | genomic extraction | TTACGGGACGGCGGTC |
| RB41:MG1655-extr_fw | 2013-07-15 | genomic extraction | ATGAAAACTACGCATACCTCCCTC |
| RB42:MG1655-extr_rv | 2013-07-15 | genomic extraction | TTAATCGTGTTGGCACAGCGTTATG |
| RB43:Bsub-extr_fw | 2013-07-15 | genomic extraction | ATGAAGATTTACGGAATTTATATGGAC |
| RB44:Bsub-extr_rv | 2013-07-15 | genomic extraction | TTATAAAAGCTCTTCGTACGAGAC |
| RB45:Sc-indC-Schr_rvG | 2013-07-15 | indigoidine exchangeable construct | CAT GGATCC GGTTTCCTGTGTGAA TTA TCAGTAGTTGGGCGTCTTG |
| RB46:indC-woPPT_rv | 2013-08-05 | indC-pSB1C3 w/o PPTase; RB21 overlap | GCACTATCAGCGTTATTA GCTAGCTCATTA TTA TTAGATTATTTTCTCAATCTCAG |
| RB47:indC-SpeI_fw | 2013-08-05 | remove cutting sites from indC-Plum | CAAGTTCCTAAACCC ACTAGT CTGGCTTAT ATTATTTATACCTCTGGTAGCAC |
| RB48:indC-EcoRI_rv | 2013-08-05 | remove cutting sites from indC-Plum | CAATACCCACC GAATTC TTTGAGCT AATTTCTGACAGACAATACC |
| RB49:indC-EcoRI_fw | 2013-08-05 | remove cutting sites from indC-Plum | AGCTCAAA GAATTC GGTGGGTATTG GGCTTTTTTGTGATC |
| RB50:indC-SpeI_rv | 2013-08-05 | remove cutting sites from indC-Plum | ATAAGCCAG ACTAGT GGGTTTAGGAACTTG GAACTTGAACTGTG |
| RB51:pSB(2/3)K3_fw | 2013-08-05 | PPTase plasmid | TAATGA GCTAGC tactagtagcggccgctgcagtc |
| RB52:pSB(2/3)K3_rv | 2013-08-05 | PPTase plasmid | CAT GGATCC GGTTTCCTGTGTGAA ctctagaagcggccgcgaattcc |
| RB53:entF-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAAACGATTATCGCCGCGGCATTC |
| RB54:entF-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT TGCCAGTTTGGCGACAGTTGACG |
| RB55:tycA1-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAATCGATTCTCGTCTCCATCTGG |
| RB56:tycA1-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT AGCAACCTGCTCGATCGTCGGGTAATTC |
| RB57:tycC6-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAACAGCAACTGGCAGCCATCTGGCAAG |
| RB58:tycC6-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT GGCCACCTTTTCGATCGTCGGATACTG |
| RB59:delH4-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAGACGCTGCTGGCCCGTATCTG |
| RB60:delH4-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT CGCCAGCTCGGCAATGCTTTGC |
| RB61:delH5-T_fw | 2013-08-07 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GCCATGGCGCTGGCCCGCATC |
| RB62:delH5-T_rv | 2013-08-07 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT CAGCAGGCCGGCAATGGTCG |
| RB63:pSB(2/3)K3_rv_korr | 2013-08-07 | PPTase plasmid | CAT GGATCC GGTTTCCTGTGTGAA CTCTAGTATGTGTGAAATTGTTATCC |
| RB64:DelC-extr_fw | 2013-08-07 | get Delftia PPTase | ATGCAGCTCGTGTCCGTGCGTGAG |
| RB65:DelC-extr_rv | 2013-08-07 | get Delftia PPTase | TCATGTCGATTCCTTGGTGCGCCAC |
| RB66:DelC-pSB2K3_fw | 2013-08-07 | DelC in PPTase-construct | CACAGGAAACC GGATCC ATGCAGCTCGTGTCCGTGCGTGAG |
| RB67:DelC-pSB2K3_rv | 2013-08-07 | DelC in PPTase-construct | CAGCGTTATTA GCTAGC TCA TCATGTCGATTCCTTGGTGCGCCAC |
| RB68:indC-SpeI_fw_corr | 2013-08-09 | indC w/o cutting sites corrected | CAAGTTCCTAAACCC ACgAGT CTGGCTTAT ATTATTTATACCTCTGGTAGCAC |
| RB69:indC-EcoRI_rv_corr | 2013-08-09 | indC w/o cutting sites corrected | CAATACCCACC GAAcTC TTTGAGCT AATTTCTGACAGACAATACC |
| RB70:indC-EcoRI_fw_corr | 2013-08-09 | indC w/o cutting sites corrected | AGCTCAAA GAgTTC GGTGGGTATTG GGCTTTTTTGTGATC |
| RB71:indC-SpeI_rv_corr | 2013-08-09 | indC w/o cutting sites corrected | ATAAGCCAG ACTcGT GGGTTTAGGAACTTG GAACTTGAACTGTG |
| RB72:bpsA-TTE_rv | 2013-08-09 | exchange T and TE-domain | GCACTATCAGCGTTATTA GCTAGC TCATTA TTCCCCCAGCAGGTATCTAATATG |
| RB73:entF-TTE_rv | 2013-08-09 | exchange T and TE-domain | GCACTATCAGCGTTATTA GCTAGC TCA TTACCTGTTTAGCGTTGCGCGAATAATCG |
| RB74:tycC6-TTE_rv | 2013-08-09 | exchange T and TE-domain | GCACTATCAGCGTTATTA GCTAGC TCA TTATTTCAGGATGAACAGTTCTTGC |
| RB75:delH5-TTE_rv | 2013-08-09 | exchange T and TE-domain | GCACTATCAGCGTTATTA GCTAGC TCATTA TCAGTCCAGCGCGTACTCCAG |
| RB76:plu2670-T_fw | 2013-08-15 | bring T-Domain in indC | TGGTGCCATTACATACAGATACT GAAACCACACTGGCTGCTATC |
| RB77:plu2670-T_rv | 2013-08-15 | bring T-Domain in indC | AGAGTCTGTCTGTTCAATCCACTT CGCAAATGCGGATAACACC |
| RB78:indC-SpeI_rv_corr2 | 2013-08-26 | indC w/o cutting sites w/o frameshift | ATAAGCCAG ACTcGT GGGTTTAGGAACTTG AACTGTG |
| RB79:indC_woPPTase_rv2 | 2013-08-29 | indC-pSB1C3 w/o PPTase; RB35 5' part | CAGCGTTATTA GCTAGC TCA TTAGATTATTTTCTCAATCTCAG |
| RB80:indCdT_3_fw | 2013-09-06 | GGCGGATATCCTATGAGTTTGAGATTGC | |
| RB81:indCdT_3_rv | 2013-09-06 | TCCATTGGCCGTCAAAGGTAATTTATCG | |
| RB82:bpsA-T3_fw | 2013-09-06 | CGATAAATTACCTTTGACGGCCAATGGA AAGATCGATGTGAAAGCACTGGCCGCTTCTGACC | |
| RB83:bpsA-T3_rv | 2013-09-06 | GCAATCTCAAACTCATAGGATATCCGCC CAGACCTGGCCAGCAGATCACAGG | |
| RB84:indCdT_4_fw | 2013-09-06 | TTTGAAGTTGCATACCAGCTTGAACAAGC | |
| RB85:bpsA-T4_rv | 2013-09-06 | GCTTGTTCAAGCTGGTATGCAACTTCAAA TGCCACGCGAGCTCCGAAGCTATATCC | |
| RB86:indC-T2_fw | 2013-09-06 | TTACAGATACTTTTTCAATCTCCTAATATAGCAG | |
| RB87:indC-T2_rv | 2013-09-06 | TACTTCCATCCAAATTTTTCCAAGC | |
| RB88:bpsA-T2_fw | 2013-09-06 | GCTTGGAAAAATTTGGATGGAAGTA CTGAGACGCGAAAATGCTAGTGTCC | |
| RB89:bpsA-T2_rv | 2013-09-06 | TTAGGAGATTGAAAAAGTATCTGTAA AGGCAGGGACACTCCCAGTCTAGCATTCAG | |
| RB90:indC-T-screen_fw | 2013-09-13 | GATCAATGCGGCCTTTAATATTCG | |
| RB91:bpsA-T-screen_fw | 2013-09-13 | GGAACTGAATGCTAGACTGGGAG | |
| RB92:entF-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGCTGGATCGCAAAGCCTTACCGTTG | |
| RB93:tycA1-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGATCGACCGCAAAGCGTTGC | |
| RB94:tycC6-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAAGTGGATCGCAAAGCTTTG | |
| RB95:delH4-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGCTGGACCGGCAGGCCCTG | |
| RB96:delH5-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGCTTGACCGTGGTGCGCTG | |
| RB97:plu2642-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAAATCGATTTCGACACATTACAAG | |
| RB98:plu2670-T3_fw | 2013-09-16 | CGATAAATTACCTTTGACGGCCAATGGA AAGCTGGACCGTCGGGCGTTACCGGCAC | |
| RB99:plu2642_pRB23-plu_fw | 2013-09-24 | plu2642 on pSB1C3 | CATACTAGAG AAAGAGGAGAAA GGTACC ATGCAATCAACTCTCCCAATAATAAAATGG |
| RB100:plu2642_pRB23-plu_rv | 2013-09-24 | plu2642 on pSB1C3 | CAGCGTTATTA GCTAGC TCA TCAACTCAAGAATCGAGCTAATTCGTTAACAC |
| RB101:plu2642_A_rv | 2013-09-24 | GTTCTGGCATCCTTCTTAATTAACAT TGCAGCGTGTTTCACTTGATCGATAAC | |
| RB102:indC_Ox_fw | 2013-09-24 | ATGTTAATTAAGAAGGATGCCAGAAC | |
| RB103:plu2642_valInd_fw | 2013-09-24 | CAGCCTGCCAGCAGCAAAACG ATGCAATCAACTCTCCCAATAATAAAATGG | |
| RB104:plu2642_valInd_rv | 2013-09-24 | CAT CGTTTTGCTGCTGGCAGGCTG | |
| RB105:tycC2_T3_fw | 2013-09-24 | CGATAAATTACCTTTGACGGCCAATGGA AAAGTGGATCGCAAGGCATTG | |
| RB106:tycC2_T1_rv | 2013-09-24 | AGAGTCTGTCTGTTCAATCCACTT GGCCAGGCCCGCGATCGTTGG | |
| RB107:tycC2_A_fw | 2013-09-24 | CATACTAGAG AAAGAGGAGAAA GGTACC ATGACGGAAGCGGAAAAACGCACACTCCTTC | |
| RB108:tycC2_A_rv | 2013-09-24 | GTTCTGGCATCCTTCTTAATTAACAT GACGACCGCTTTGACCGCTTCATG | |
| RB109:tycC2_valInd_fw | 2013-09-24 | CAGCCTGCCAGCAGCAAAACG ATGACGGAAGCGGAAAAACGCACACTCCTTC | |
| RB110:plu2642_TE_fw | AGTGGATTGAACAGACAGACTCT AAAATCGTTGAAGGTGAAGTAACCAG |
Plasmids
Delftibactin
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
| pIK1 | 2013-08-12 | Methylmalonyl-CoA pathway with permeability device and sfp | pSB3C5-lacP-BBa_B0034-BBa_I746200-BBa_B0029-pcc-acca2-BBa_B0030-sfp | 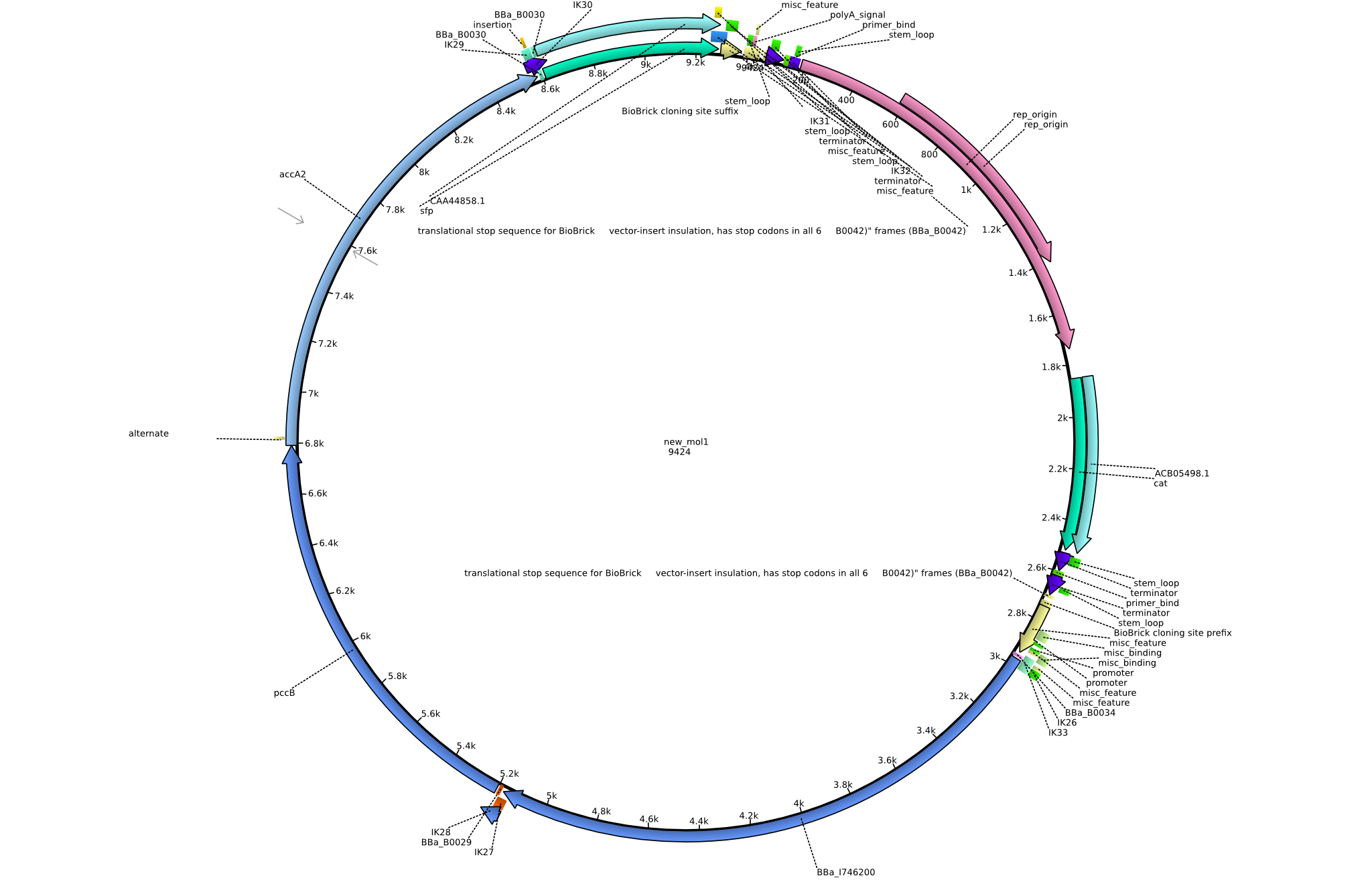 | pIK1 |
| pIK2 | 2013-08-12 | Methylmalonyl-CoA pathway with sfp | pSB3C5-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp | 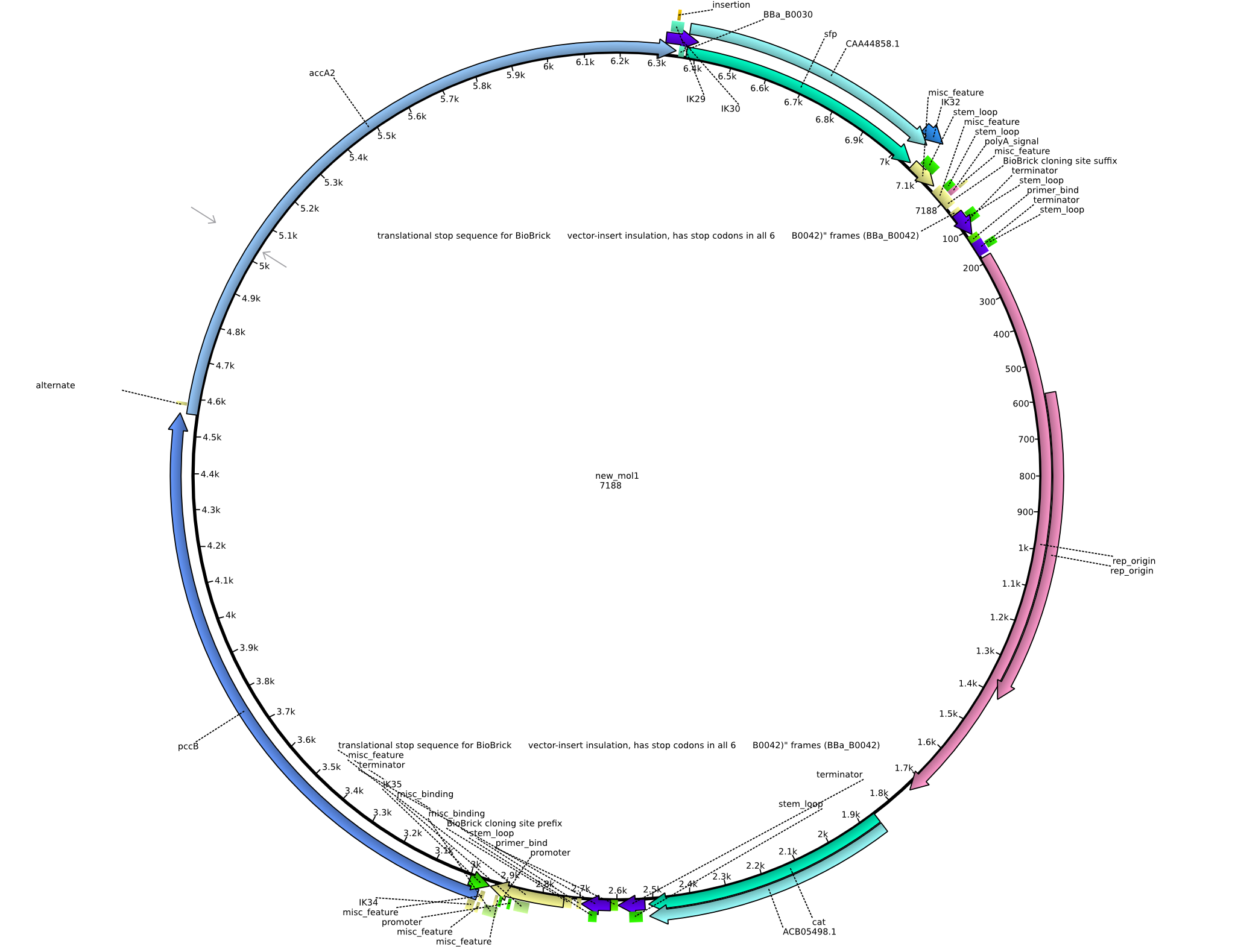 | pIK2 |
| pIK7 | 2013-08-28 | Methylmalonyl-CoA pathway with sfp | pSB3K3-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp | 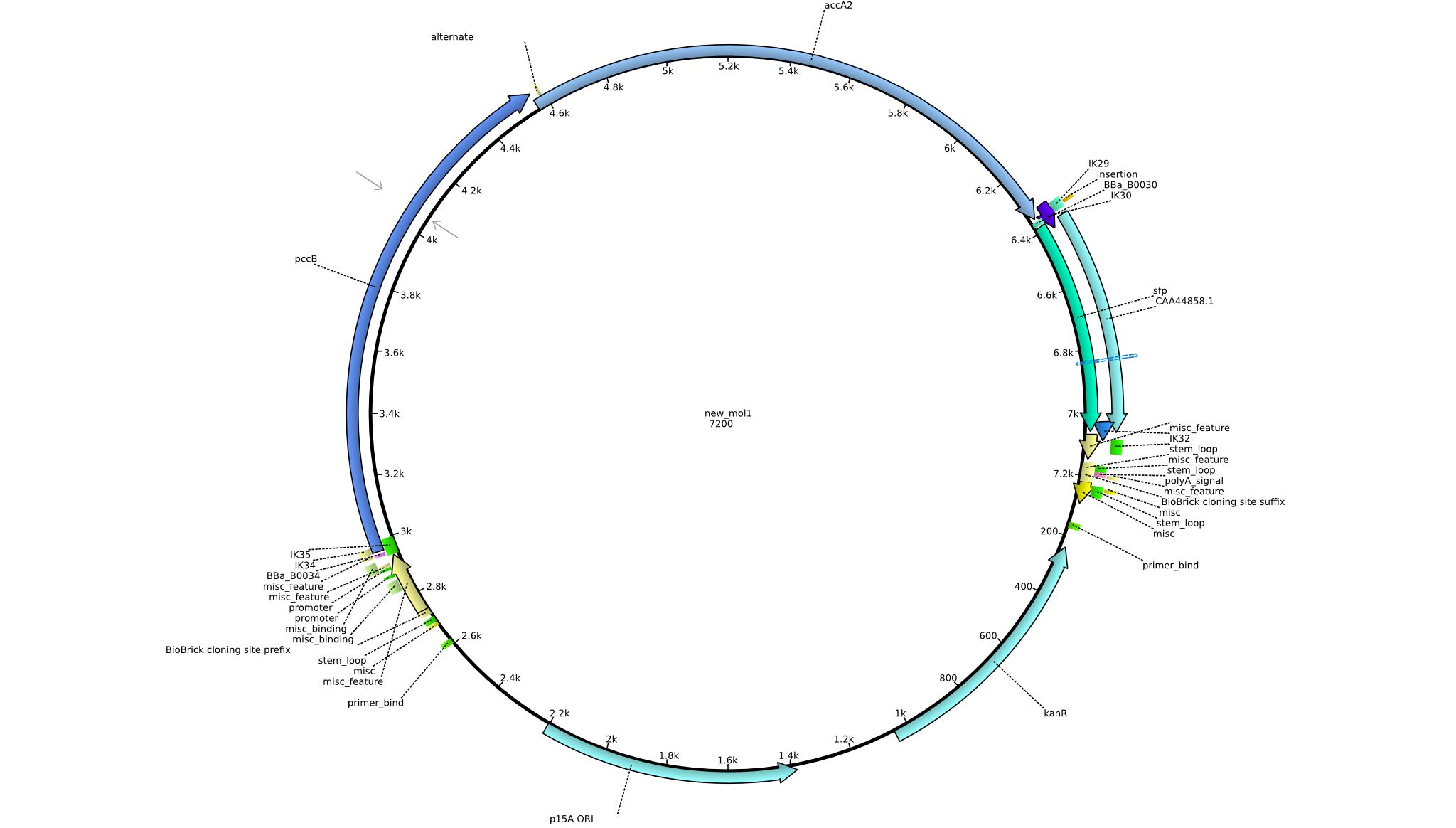 | pIK7 |
| pIK8 | 2013-09-12 | Methylmalonyl-CoA pathway with sfp and the permeability device behind a weak promoter | pSB3C5-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp-BBa_B0010-BBa_B0030-BBa_I746200-BBa_B0012 | 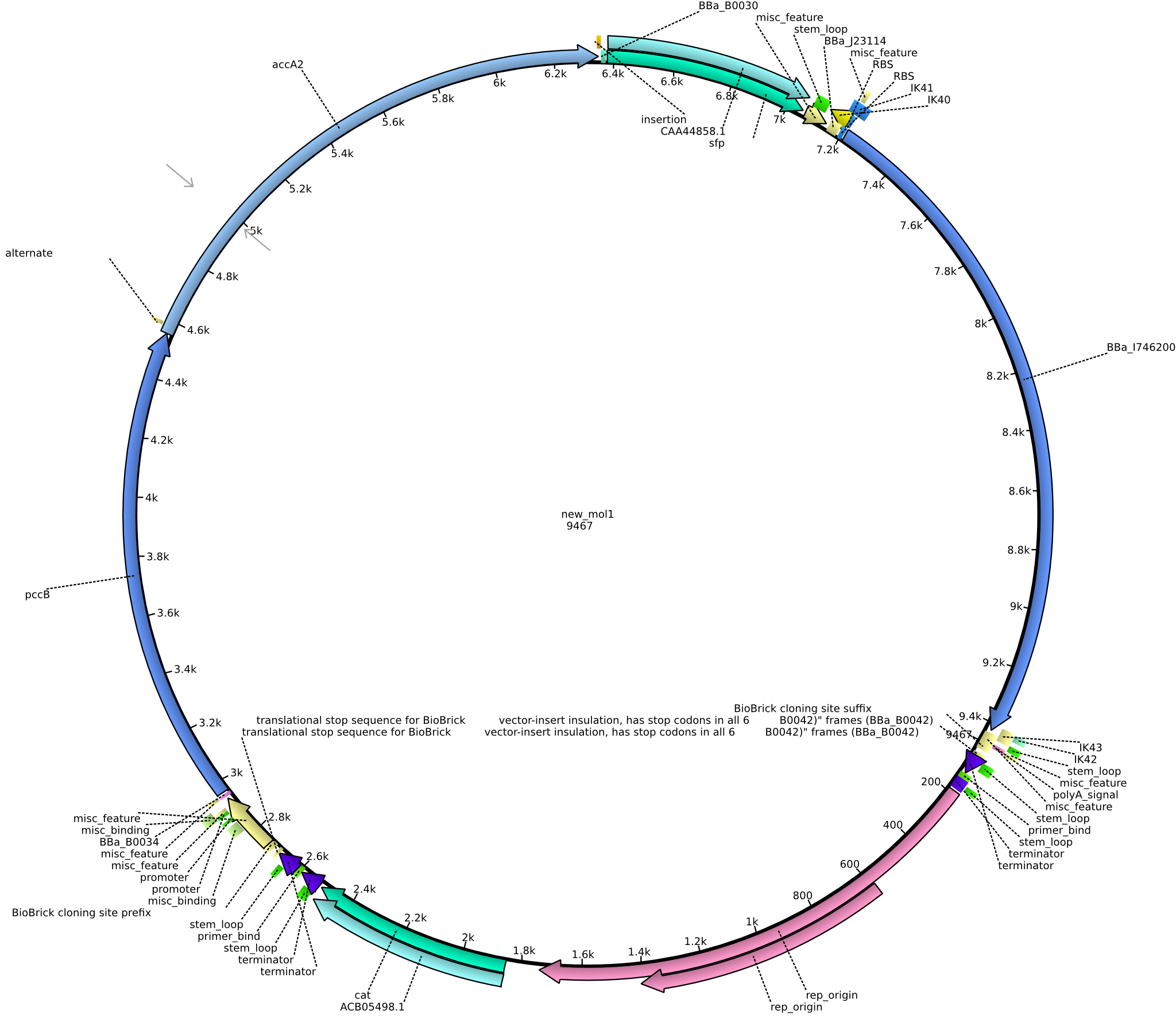 | pIK8 |
| pIK9 | 2013-09-18 | Methylmalonyl-CoA pathway with sfp and the permeability device behind a weak promoter | pSB3K3-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp-BBa_B0010-BBa_B0030-BBa_I746200-BBa_B0012 | 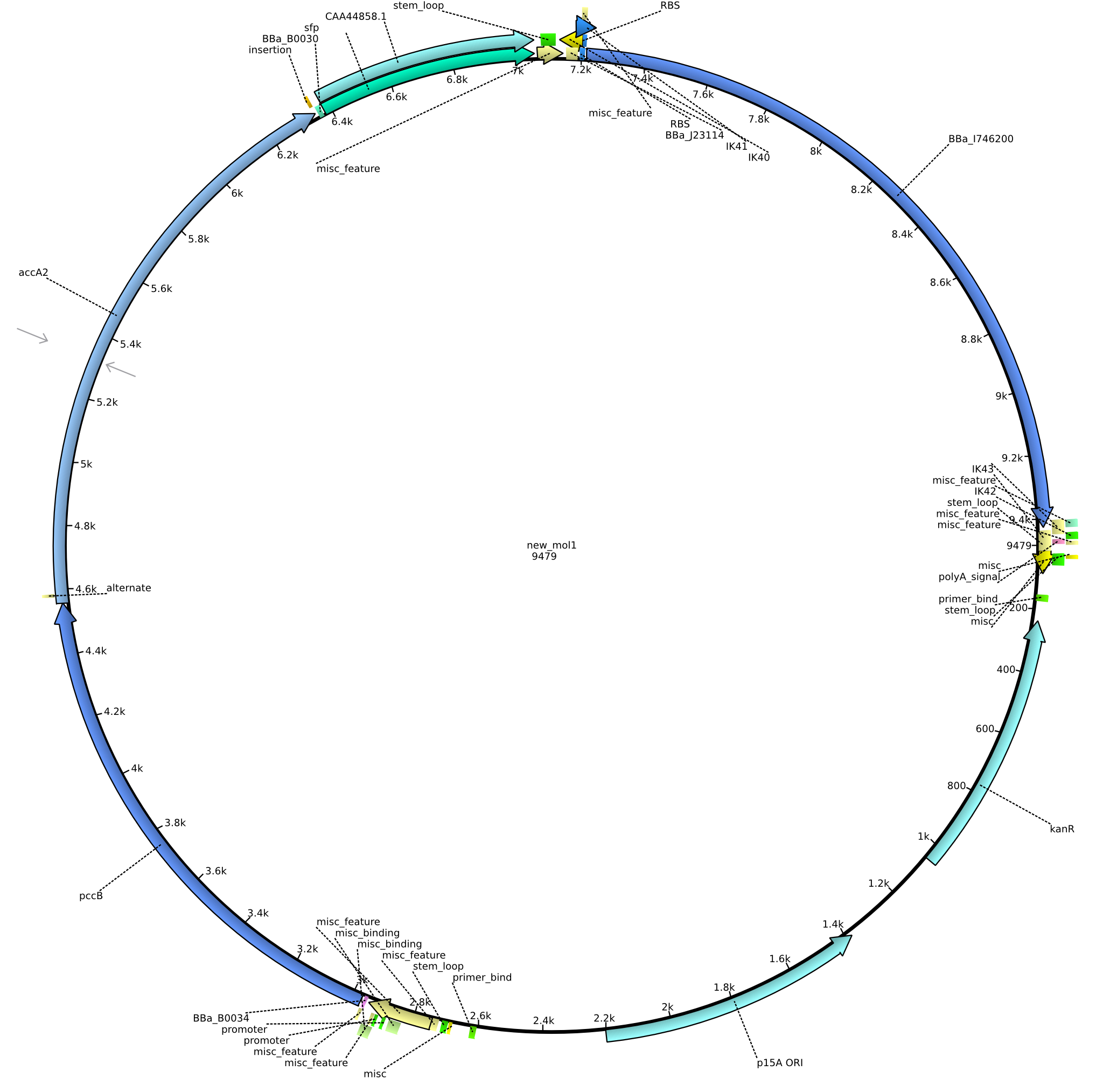 | pIK9 |
| pIK10 | 2013-09-18 | Methylmalonyl-CoA pathway with sfp and the permeability device behind a weak promoter, for submission | pSB1C3-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp-BBa_B0010-BBa_B0030-BBa_I746200-BBa_B0012 | 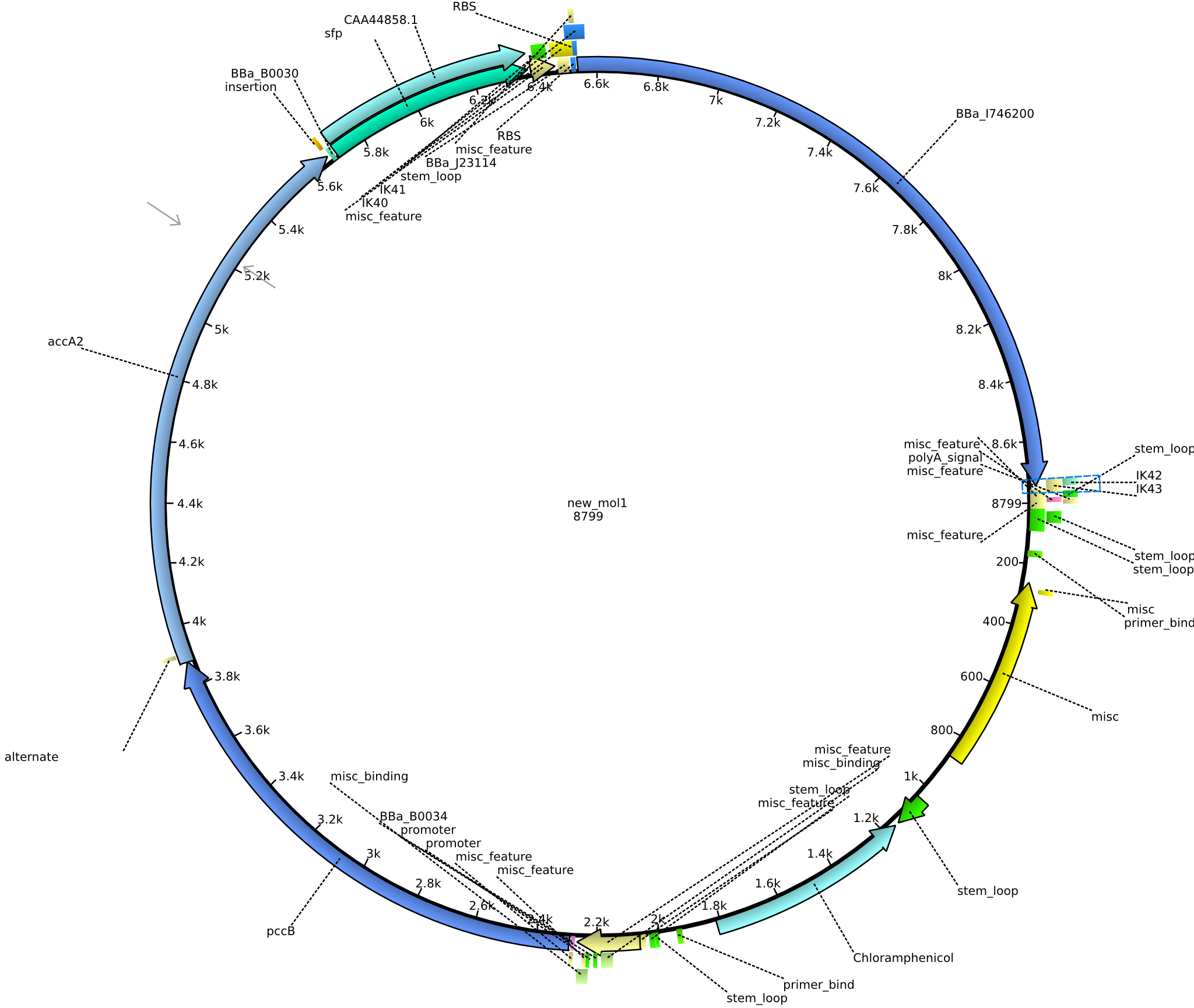 | pIK10 |
| pIK11 | 2013-09-18 | Methylmalonyl-CoA pathway with sfp, for submission | pSB1C3-lacP-BBa_B0034-pcc-acca2-BBa_B0030-sfp | 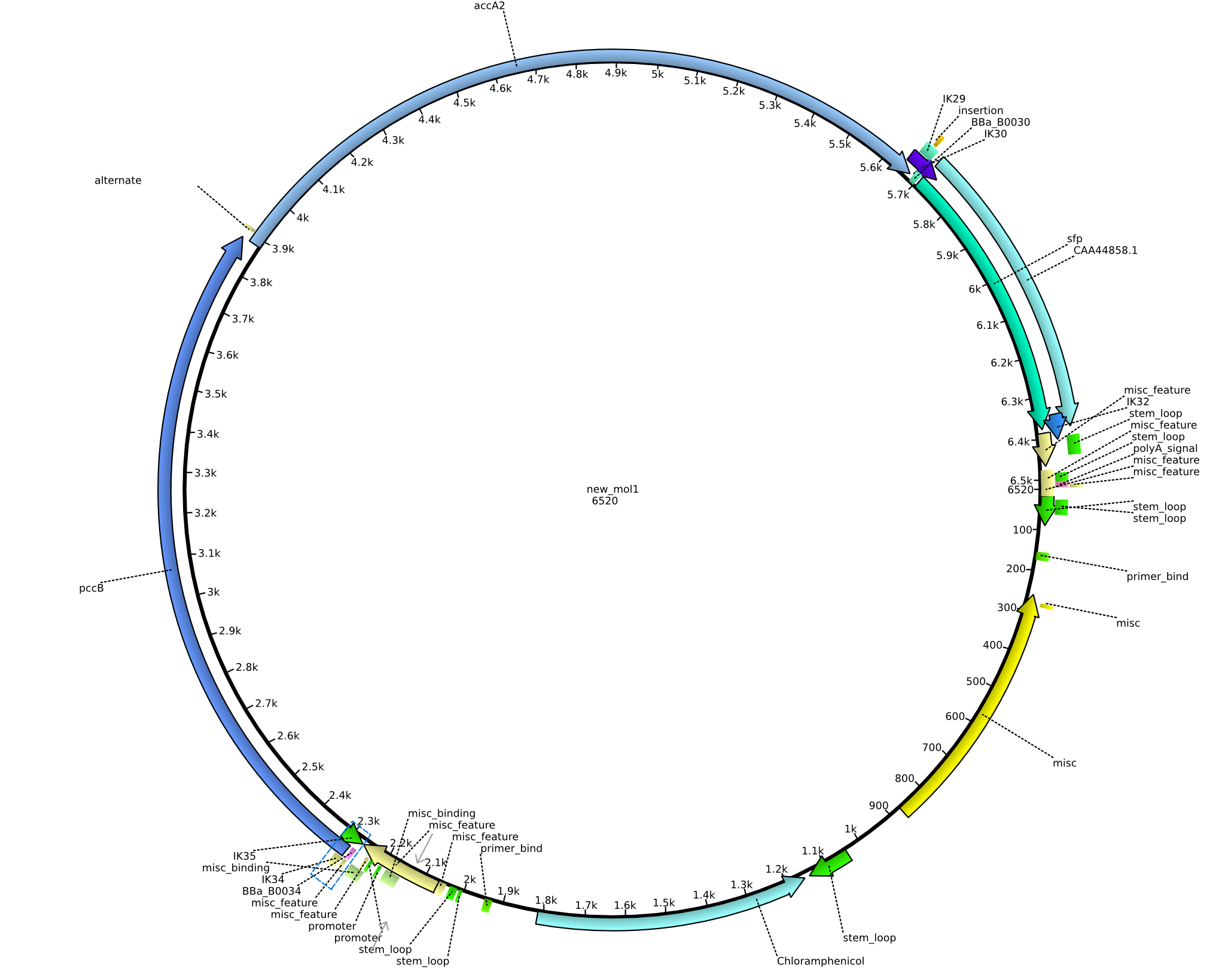 | pIK11 |
| pHM01 | 2013-04-29 | DelH construct assembled by Restriction Ligation Strategy | pSB6A1-BBa_K206000(AraC)-DelH-BBa_I732019(lacZ)-B0015(stop-codon) | 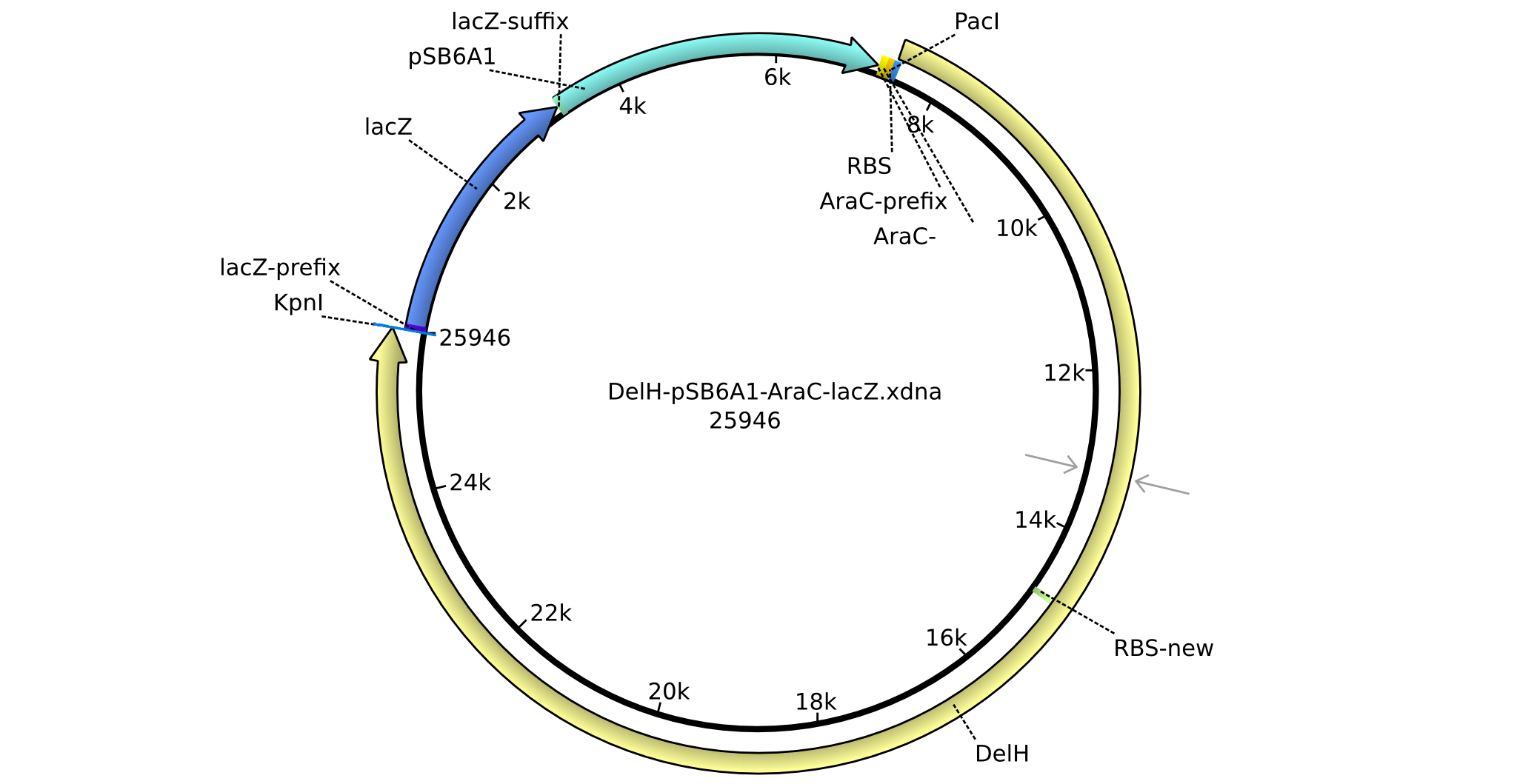 | pHM01 |
| pHM02 | 2013-07-08 | DelH construct assembled by Gibson Assemby | pSB6A1-R0010(lacI)-B0035(RBS)-DelH-B0034(RBS)-E1010(mRFP)-B0015(stop) | 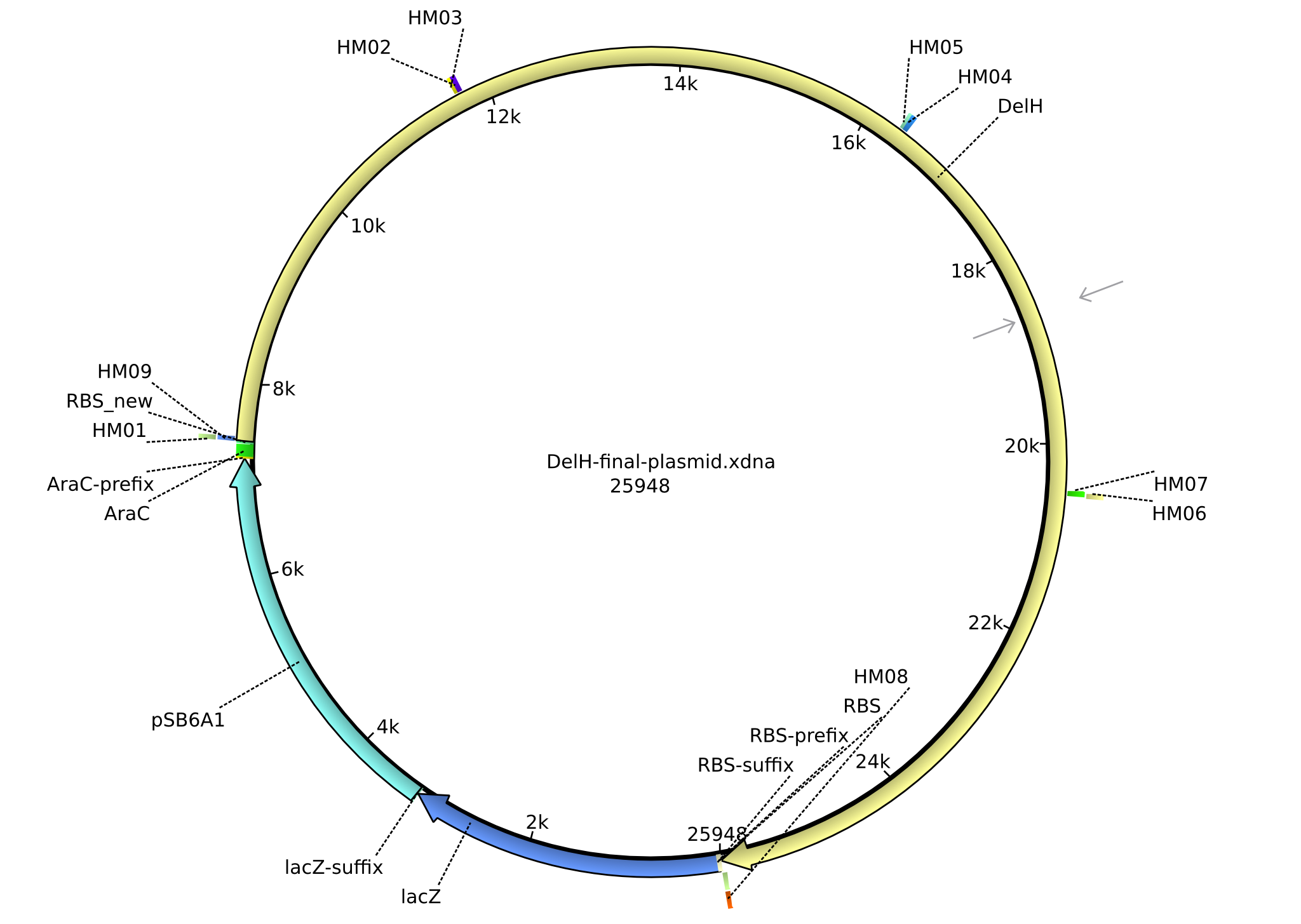 | pHM02 |
| pHM03 | 2013-04-29 | DelH construct assembled by Gibson Assemby for existing backbone | pSB6A1-R0010(lacI)-B0034(RBS)-DelH-B0035(RBS)-E1010(mRFP)-B0015(stop) | 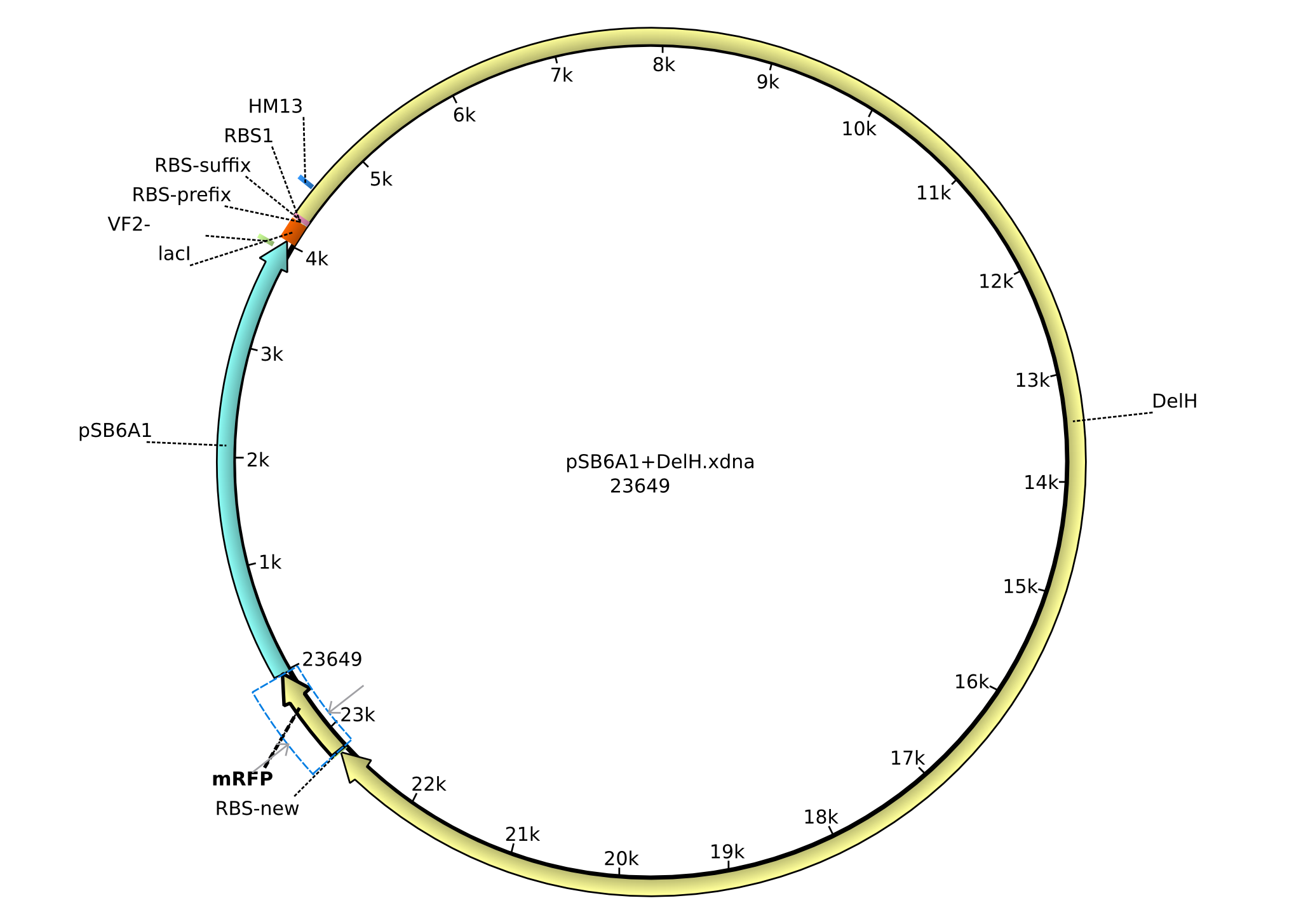 | pHM03 |
| pHM04 | 2013-08-12 | DelH construct assembled by Gibson Assemby without mRFP | pSB6A1-R0010(lacI)-B0034(RBS)-DelH-B0015(stop) | 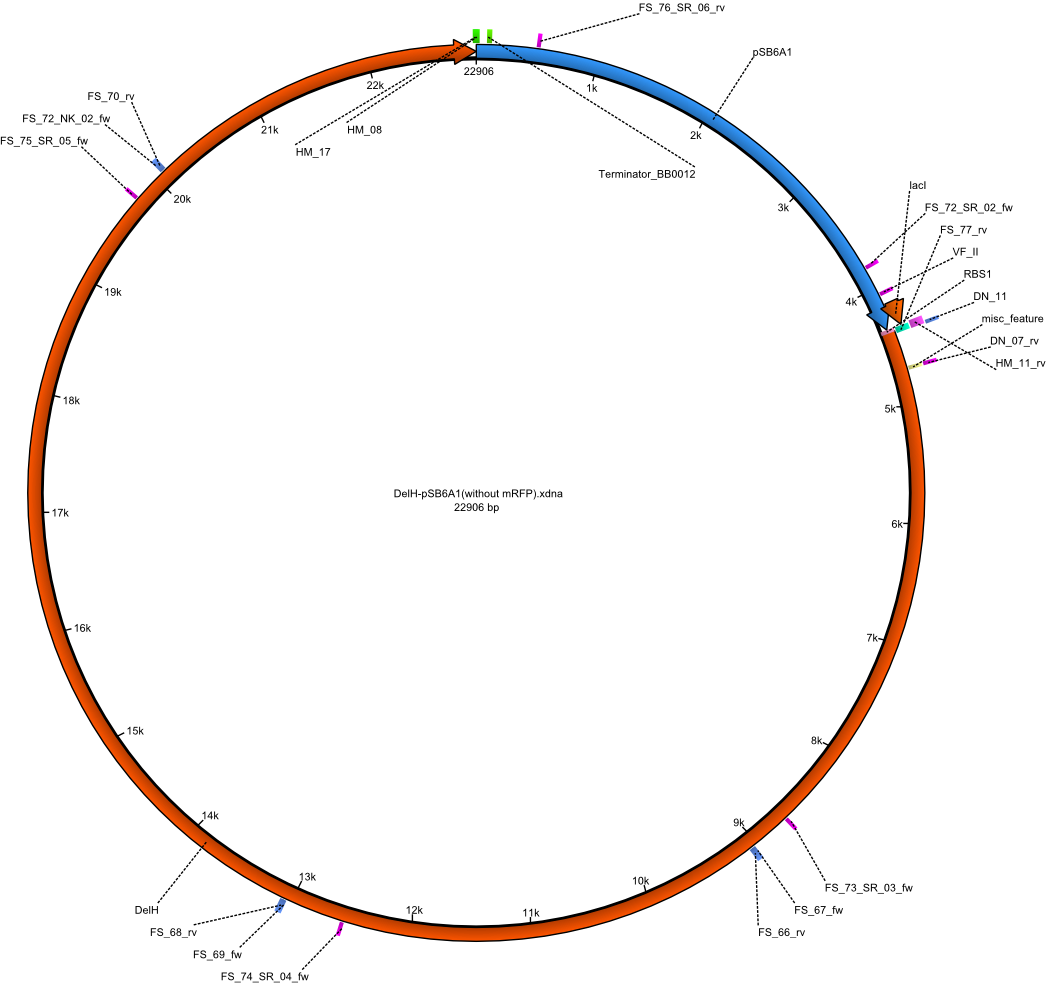 | pHM04 |
| pHM05 | 2013-08-12 | DelH construct assembled by Gibson Assemby with tetracycline resistance, without mRFP | pSB6A1-R0010(lacI)-DelH-B0015(stop)-pSB1T3(tetR) | 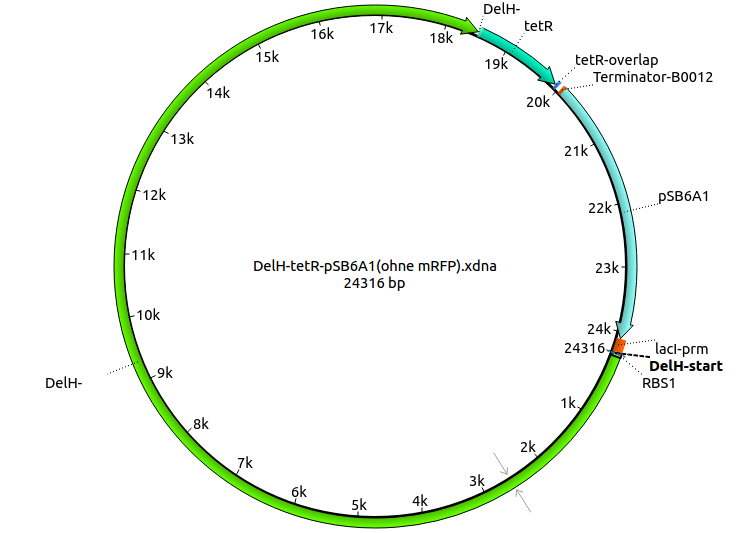 | pHM05 |
| pFSN | 2013-06-28 | DelRest, Delftibactin cluster from D. Acidovorans | pSB4K5-lacZ-BBa_B0035-DelA-DelB-DelC-DelE-DelF-DelG-DelO-DelP-DelL-BBa_B0034 | 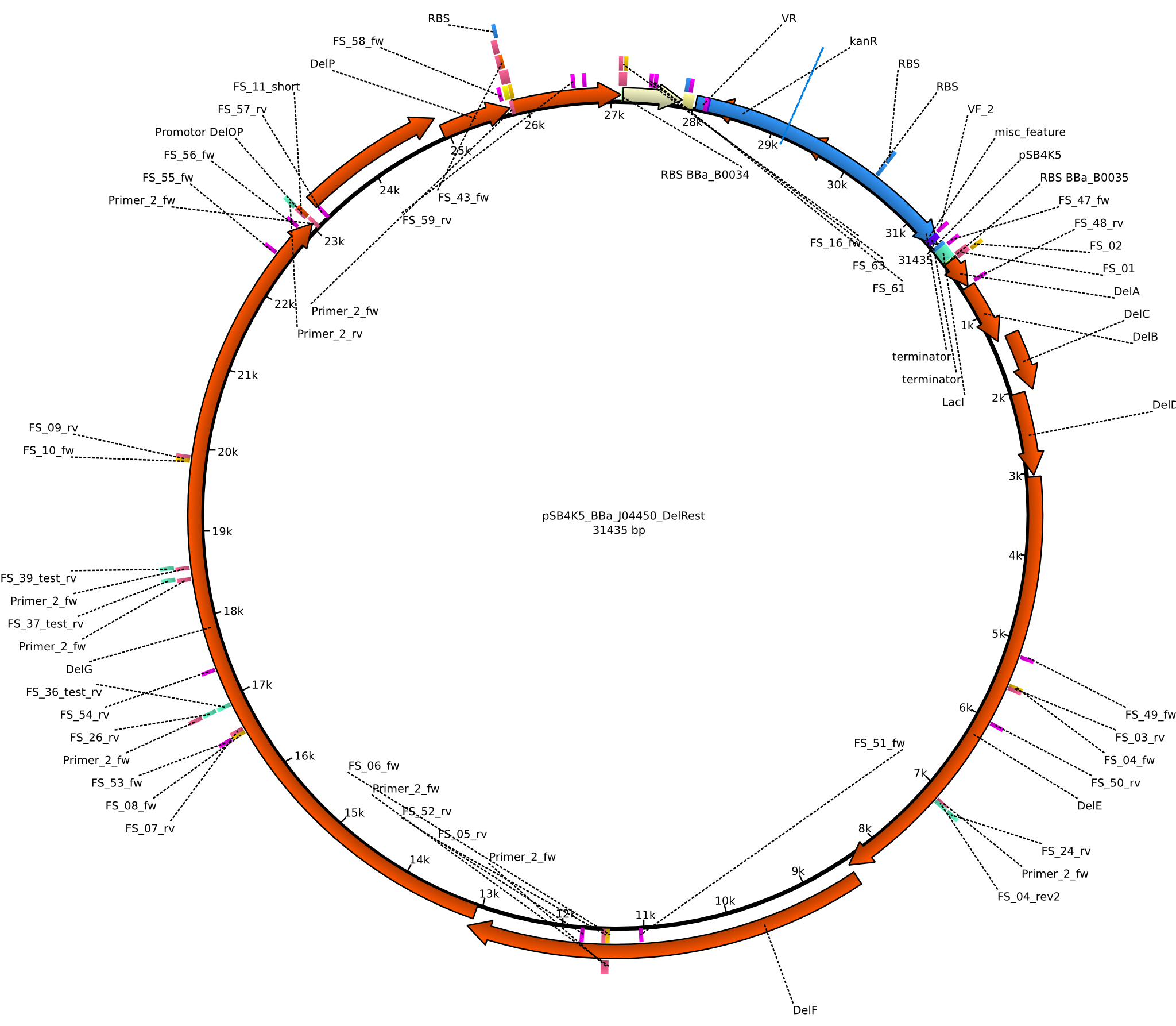 | pFSN |
| pFS_02 | 2013-09-26 | DelH construct assembled by Gibson Assemby with weak Promotor, weak RBS | pSB6A1-BBa_J23114-BBa_B0032-DelH | 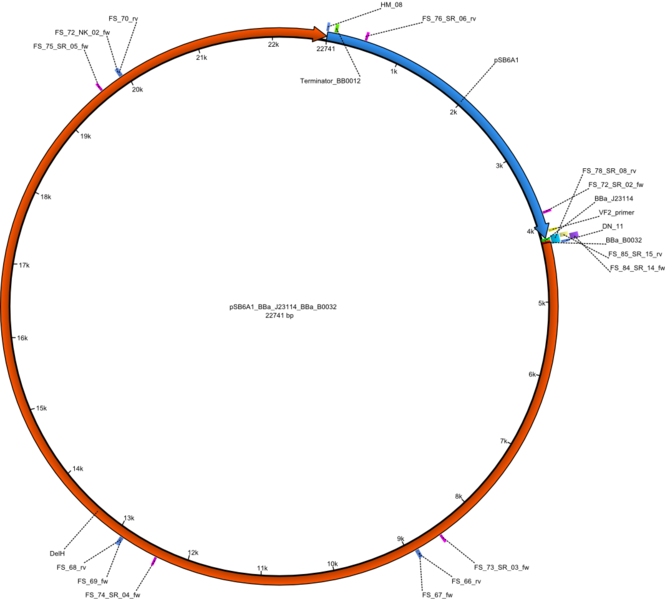 | pFS_02 |
| pFS_03 | 2013-09-26 | Helper plasmid for DelH restriction cloning, DelH, flanked by KpnI and BamHI site, without promotor, | pSB4K5-DelH | 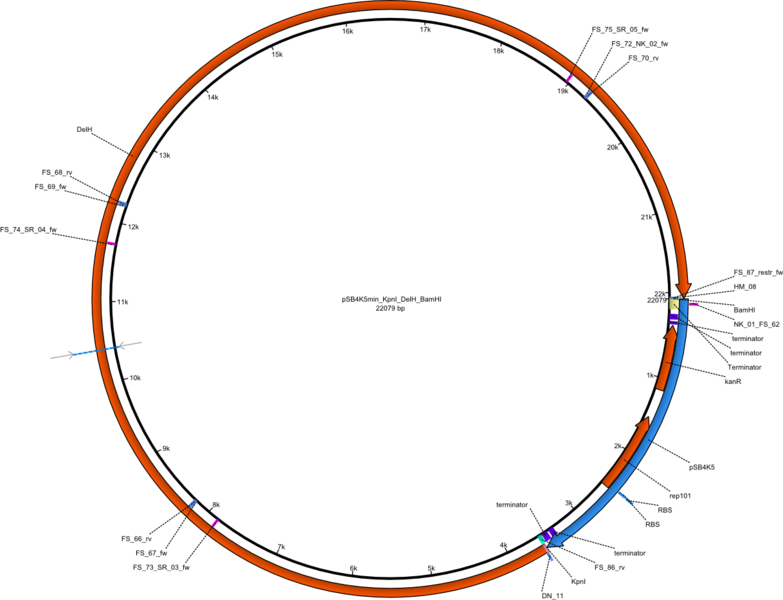 | pFS_03 |
| pFS_04 | 2013-09-26 | Target plasmid for DelH restriction cloning, ccdB cassette, flanked by KpnI and BamHI site | pSB6A1-BBa_J23114-BBa_B0032-ccdBcassette |  | pFS_04 |
| pFS_05 | 2013-09-26 | DelH construct assembled by restriction cloning using pFS_03 and pFS-04 DelH, flanked by KpnI and BamHI site, under weak promotor, | pSB6A1-BBa_J23114-BBa_B0032-DelH |  | pFS_05 |
Module Shuffling
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
| pIK03 | 2013-mm-dd | NRPS for Pro-Leu | pSB1C3+TycB1dComdC+TycC6 | 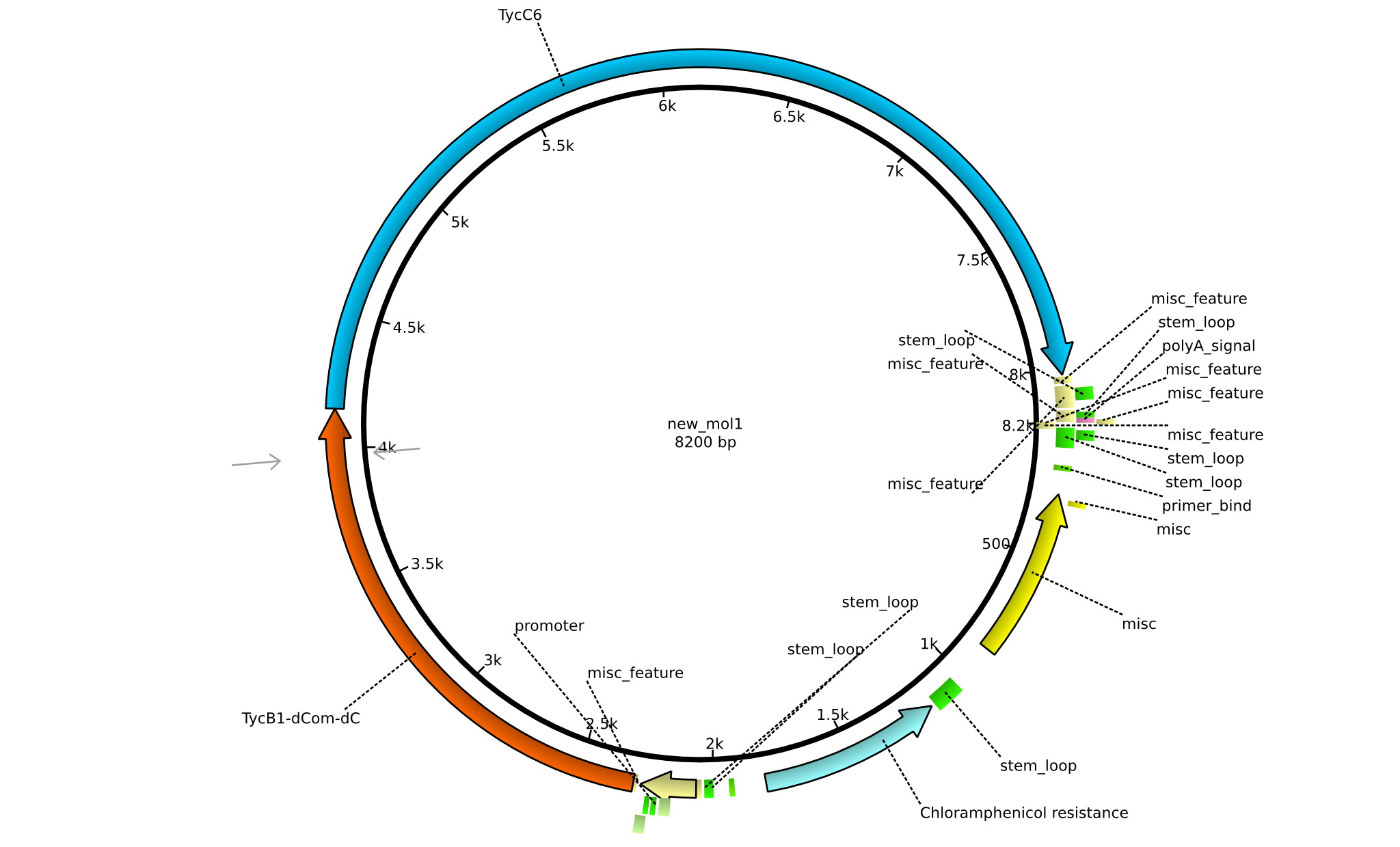 | pIK03 |
| pIK04 | 2013-mm-dd | NRPS for Phe-Orn-Leu | pSB1C3+TycAdCom+TycC5-TycC6 | 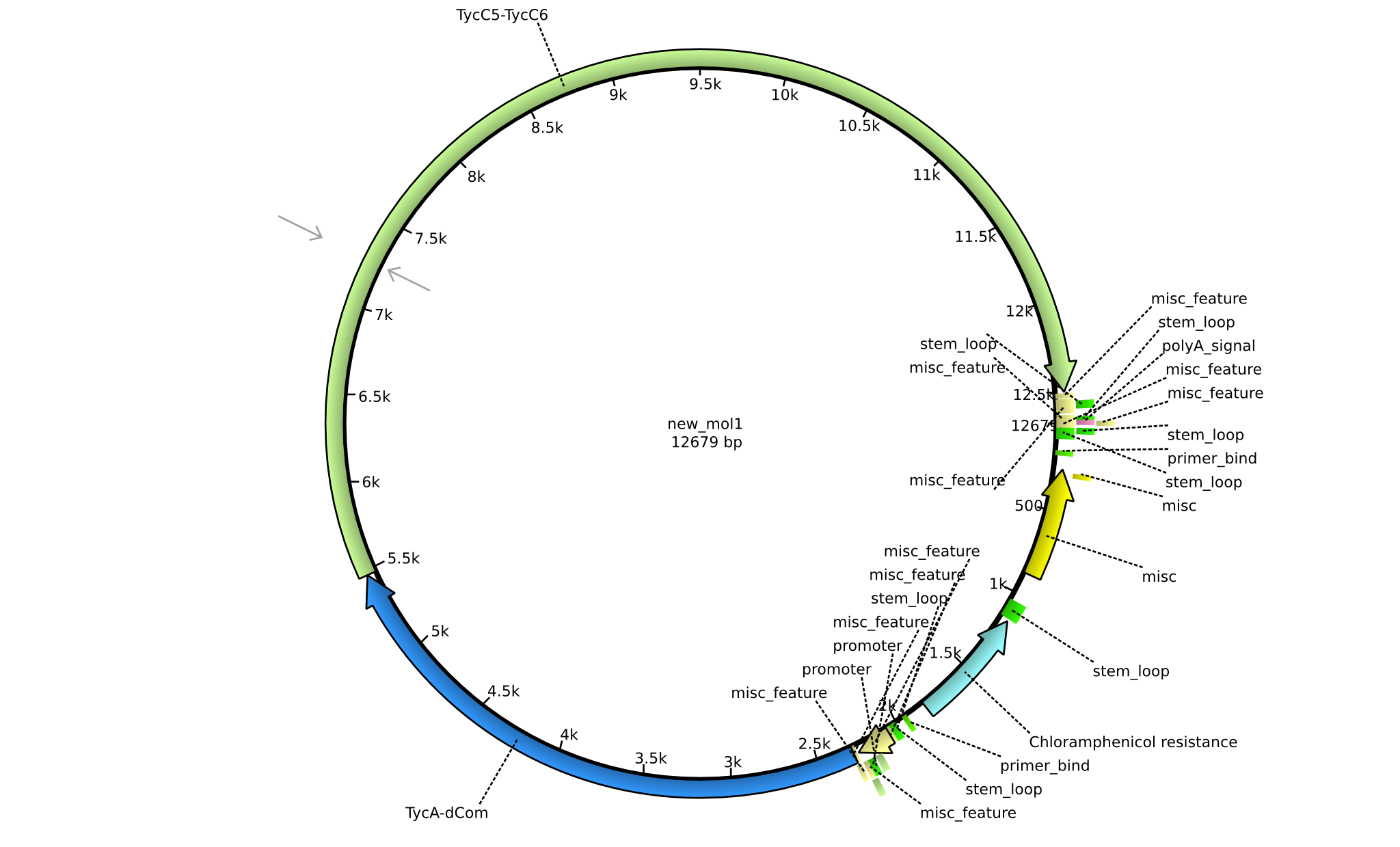 | pIK04 |
| pIK05 | 2013-mm-dd | NRPS for Pro-Orn-Leu | pSB1C3+TycB1dComdC+TycC5-TycC6 | 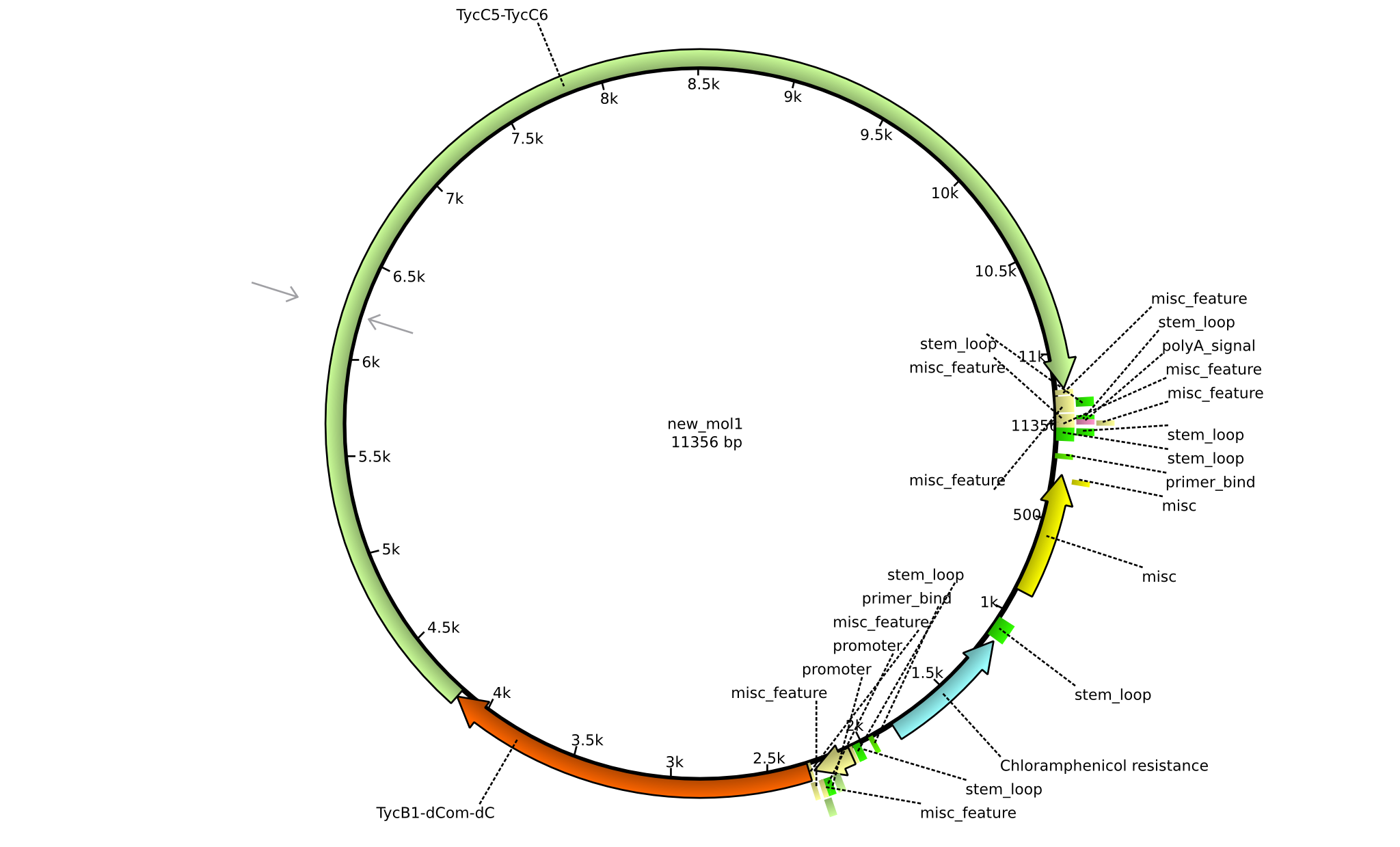 | pIK05 |
| pPW01 | 2013-mm-dd | NRPS for Orn-Pro-Phe-Leu | pSB1C3+TycC5+TycB1dCom-C(TycB2)+TycAdCom+TycC6 | 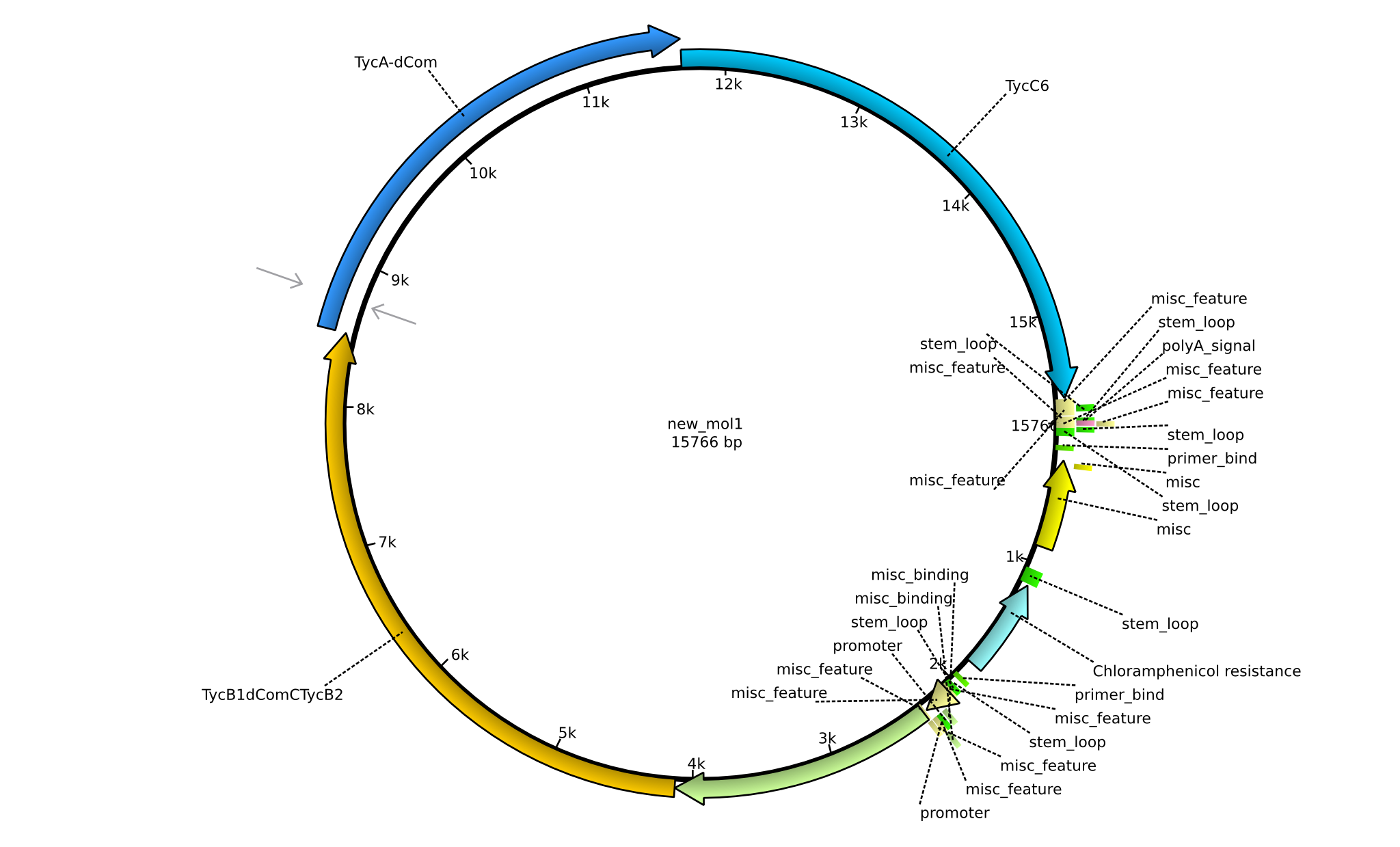 | pPW01 |
| pPW02 | 2013-mm-dd | NRPS for Orn-Pro-Phe-Leu | pSB1C3+TycC5+TycB1dCom-C(TycB2)+TycAdE+TycC6 | 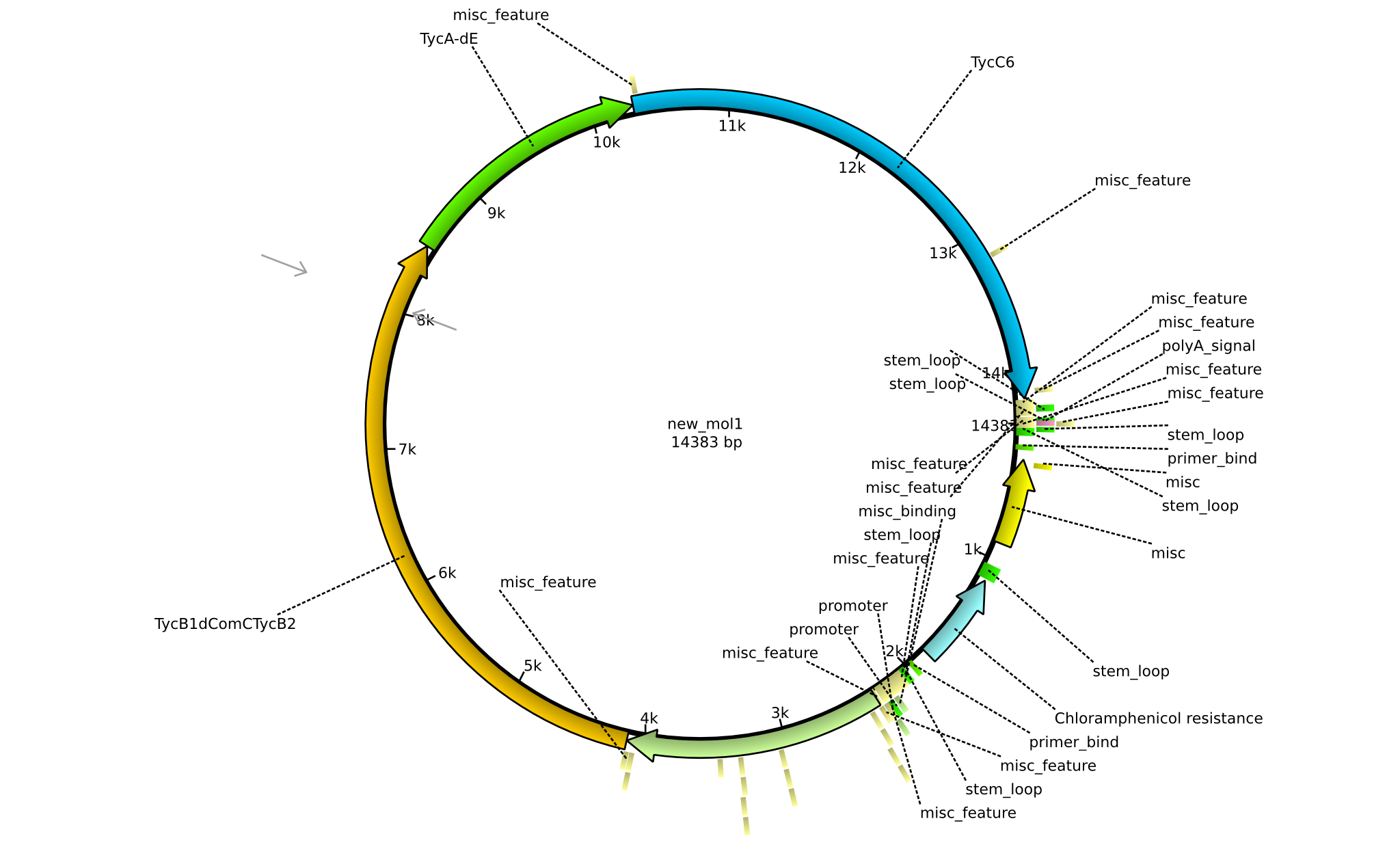 | pPW02 |
| pPW03 | 2013-08-18 | NRPS for Phe-Indigoidine | pSB1C3+TycA+C(TycC2)+IndC | 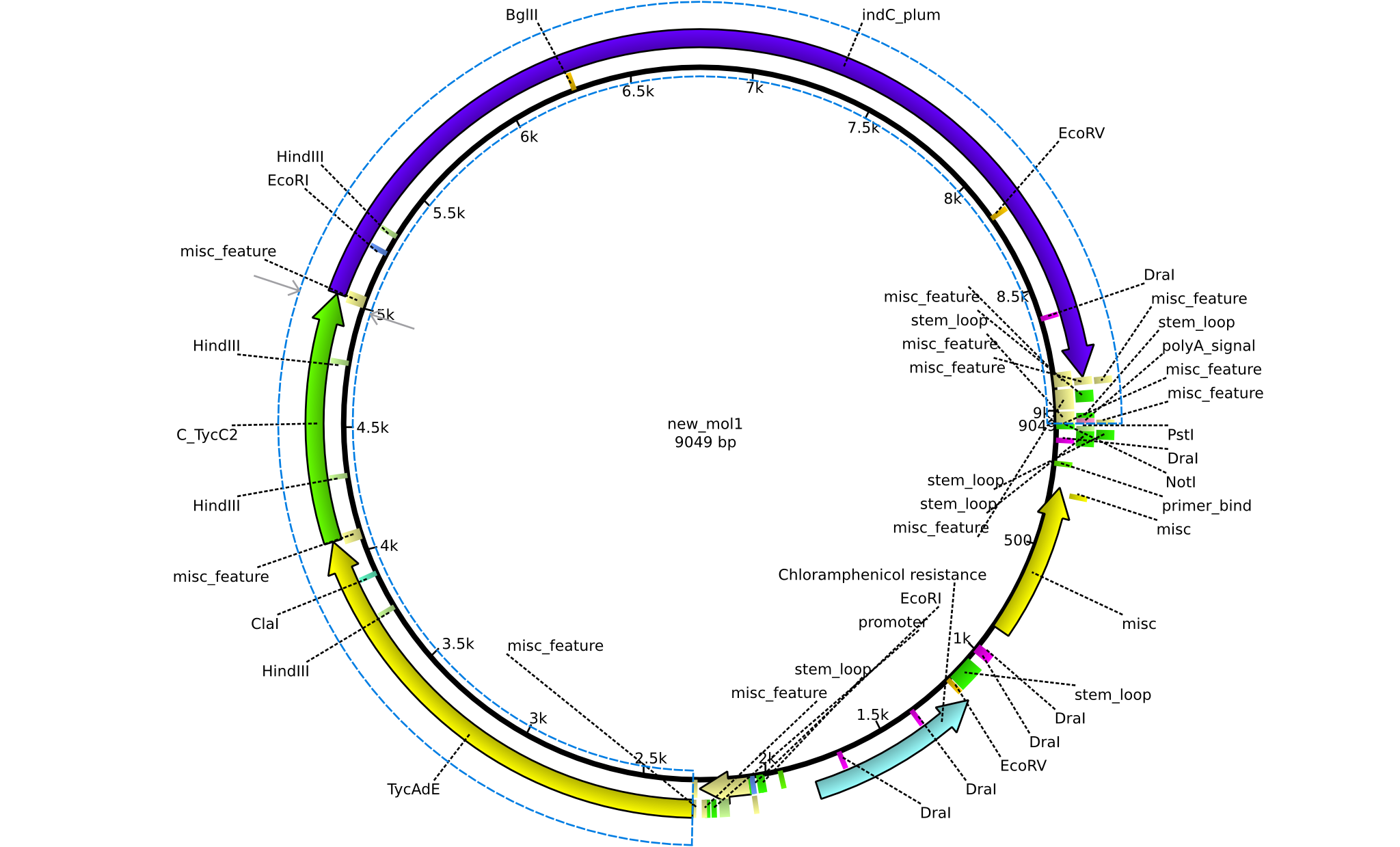 | pPW03 |
| pPW04 | 2013-08-18 | NRPS for Asn-Indigoidine | pSB1C3+TycC1-C(TycC2)+IndC | 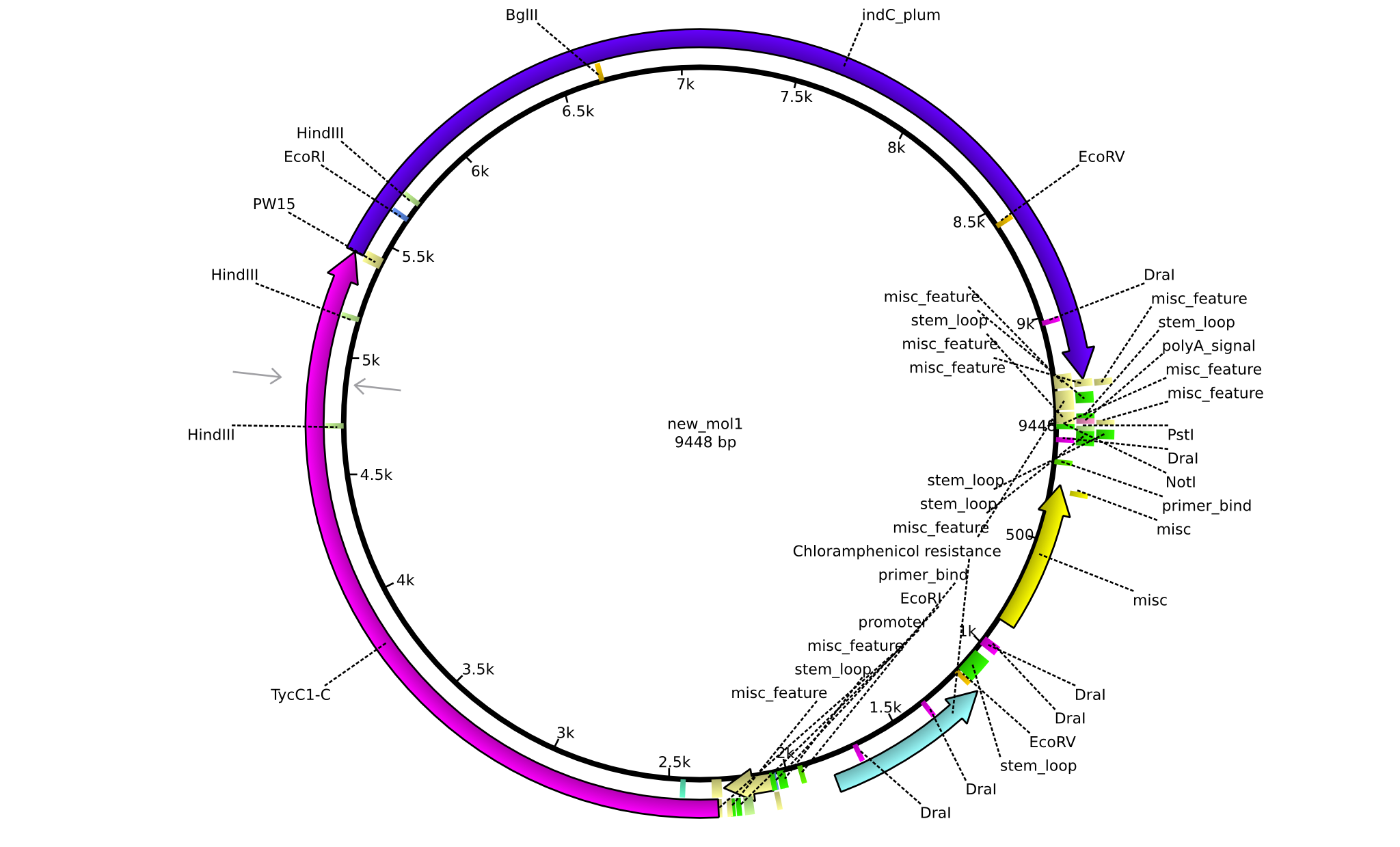 | pPW04 |
| pPW05 | 2013-08-18 | NRPS for Val-Indigoidine | pSB1C3+TycC4+C(TycC2)+IndC | 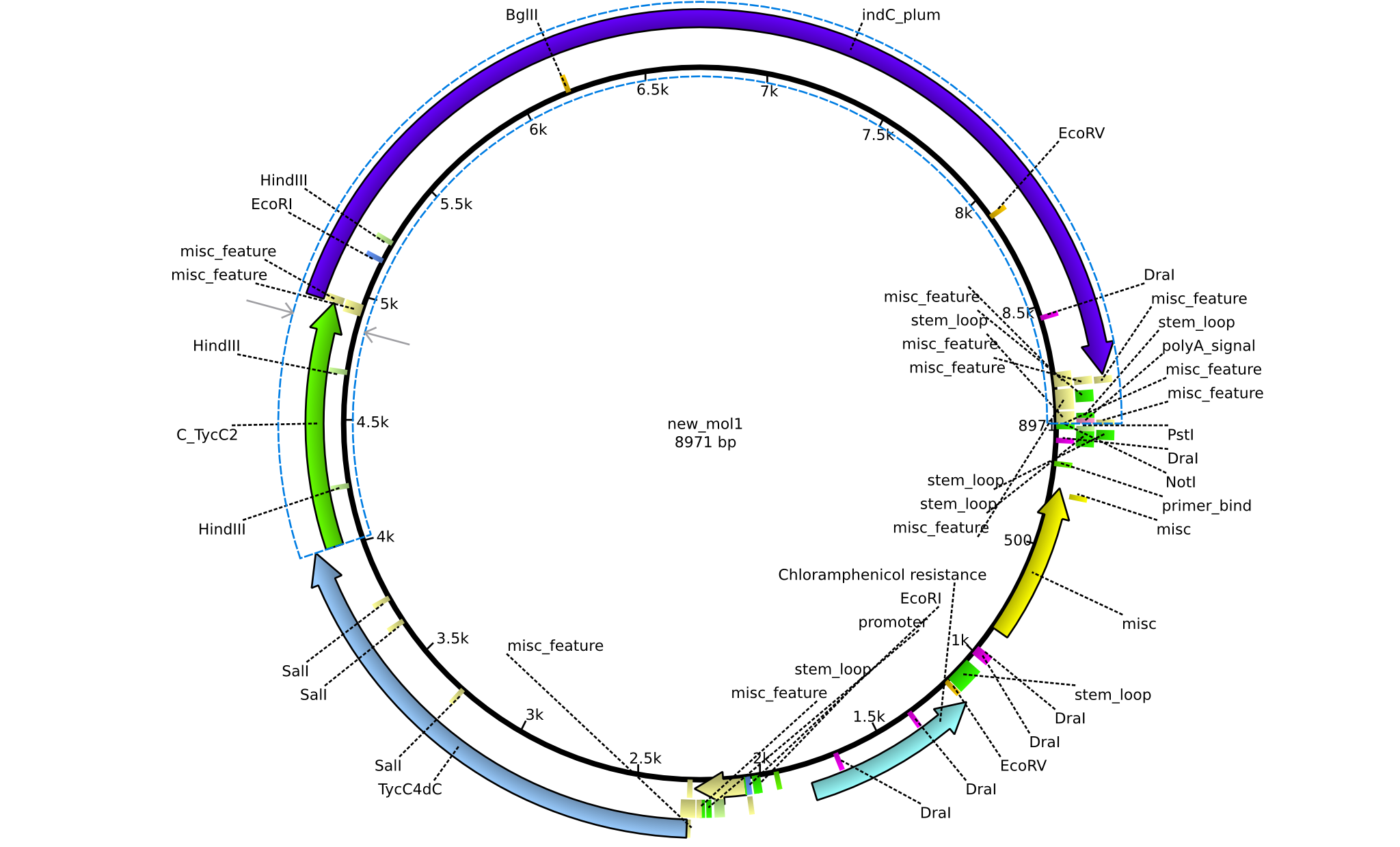 | pPW05 |
| pPW06 | 2013-09-07 | NRPS for Orn-Val-Indigoidine | pSB1C3+TycC5dC+C(TycC4)+TycC4dC+C(TycC2)+indC | 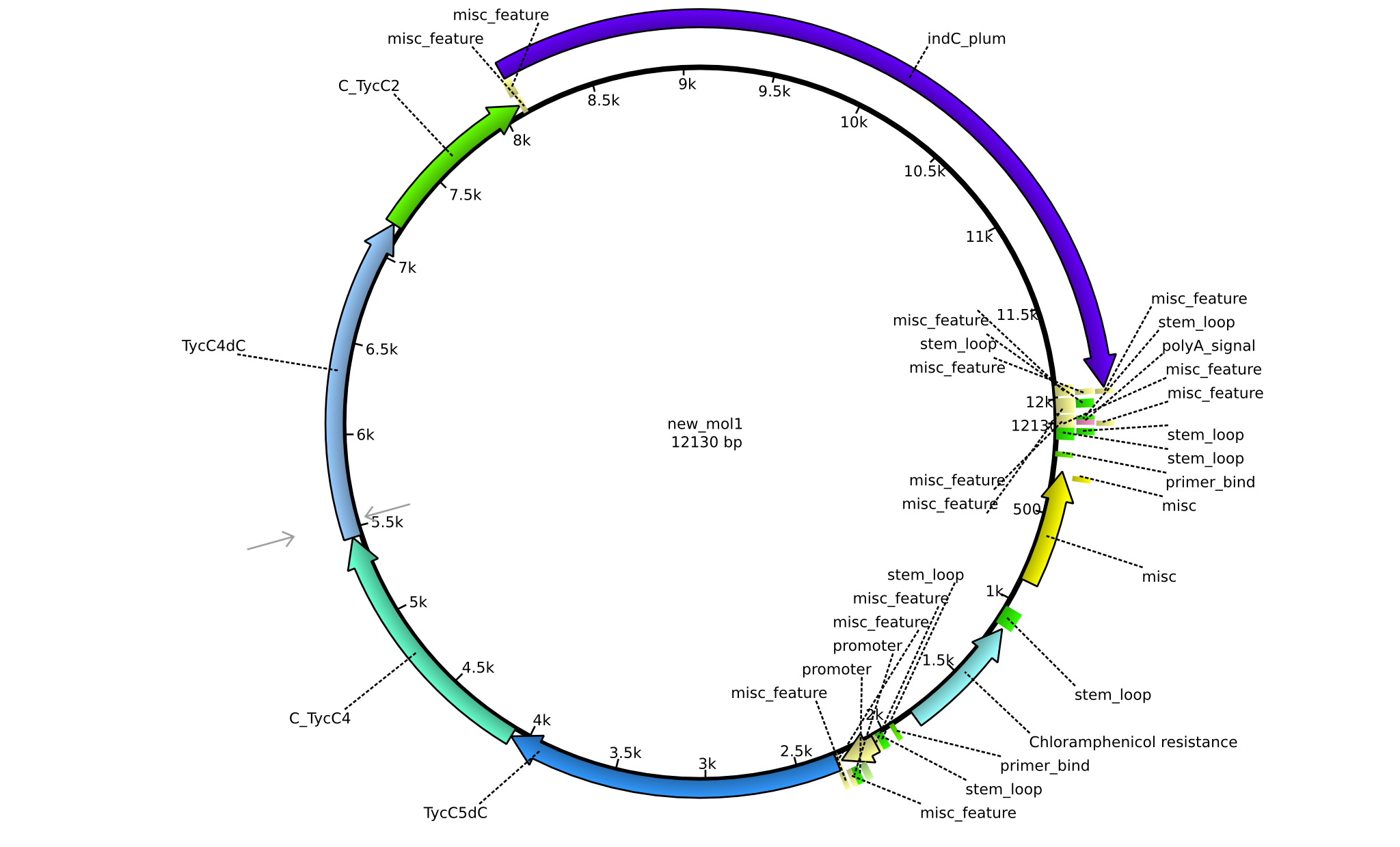 | pPW06 |
| pPW07 | 2013-09-07 | NRPS for Orn-Val-Val-Indigoidine | pSB1C3+TycC5dC+TycC4+C(TycC4)+TycC4dC+C(TycC2)+indC | 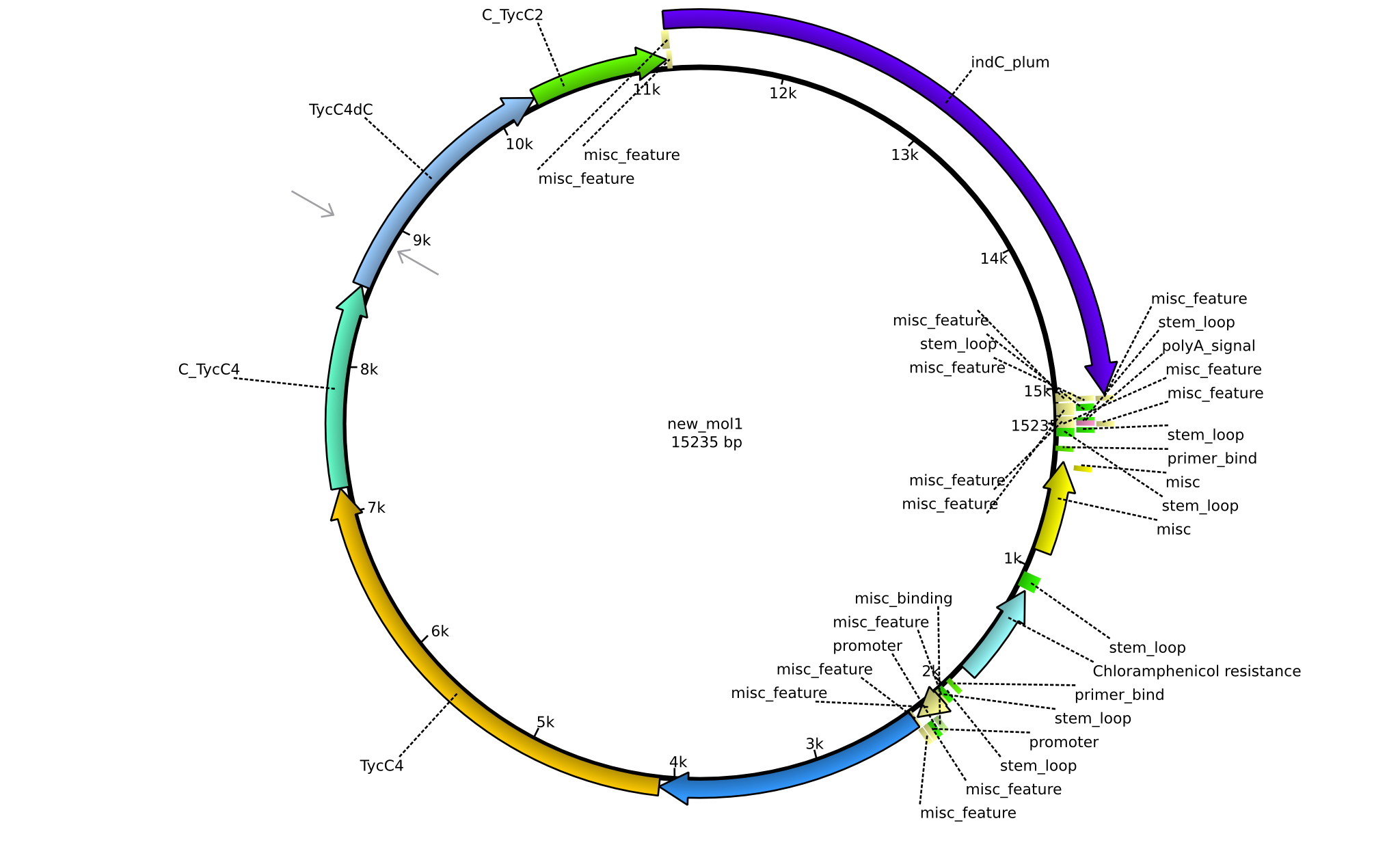 | pPW07 |
| pPW08 | 2013-09-07 | NRPS for Val-Val-Indigoidine | pSB1C3+TycC4dC+C(TycC4)+TycC4dC+C(TycC2)+indC | 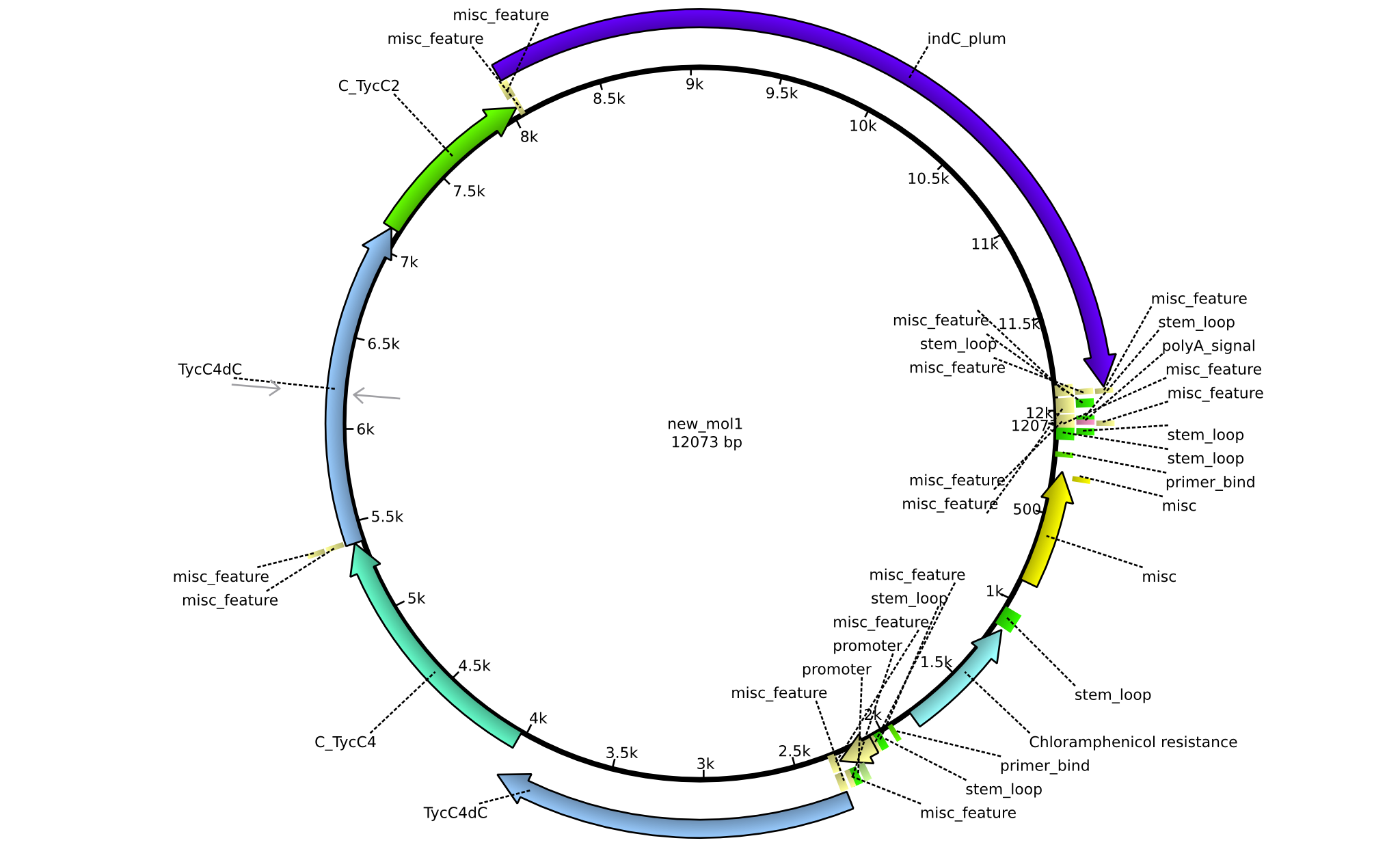 | pPW08 |
| pPW09 | 2013-09-07 | NRPS for Pro-Leu-Indigoidine | pSB1C3+TycB1dComdC+TycC6dTE+C(TycC2)+indC | 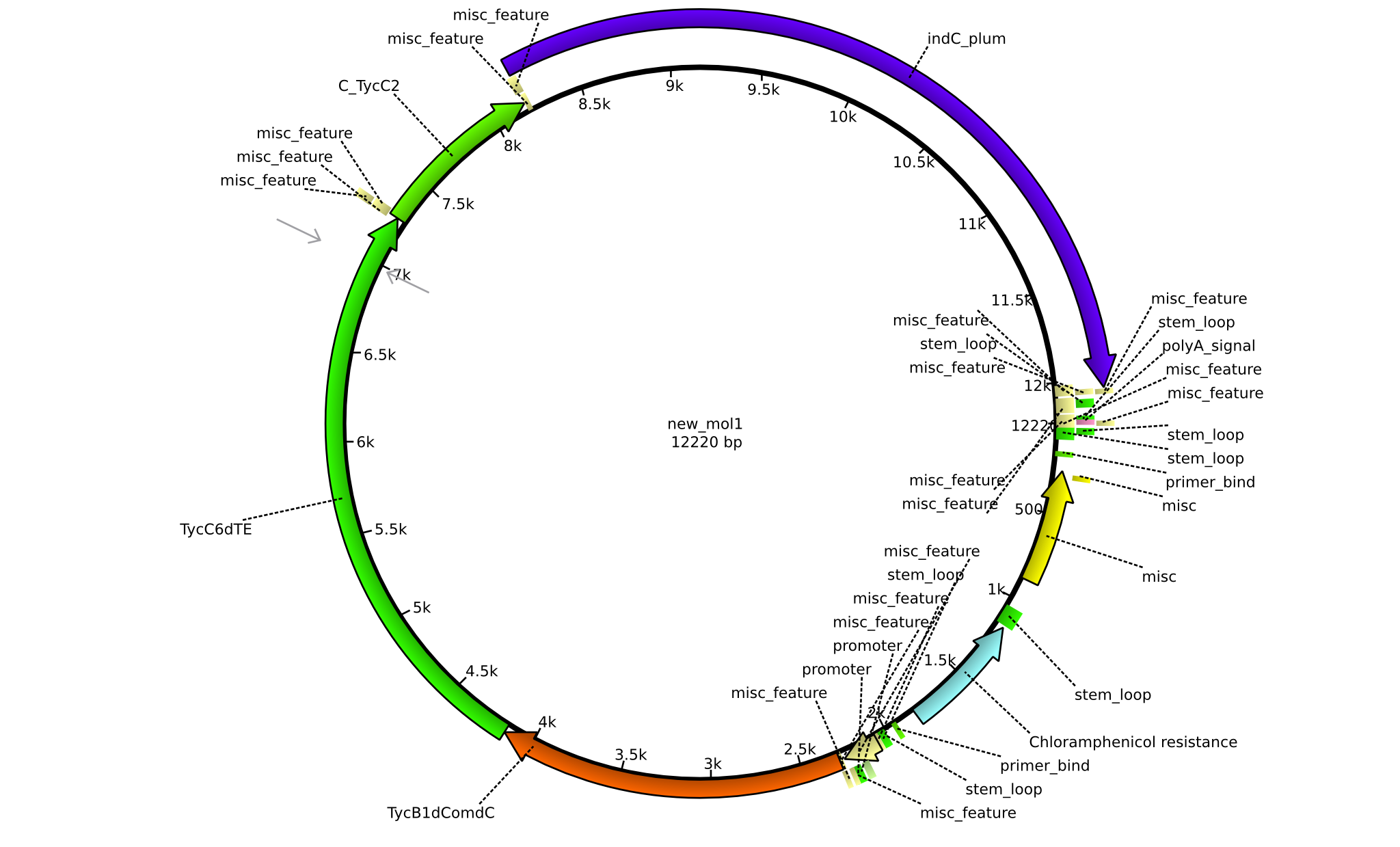 | pPW09 |
| pPW10 | 2013-09-07 | NRPS for Pro-Leu-Val-Indigoidine | pSB1C3+TycB1dComdC+TycC6dTE+C(TycC4)+TycC4dC+C(TycC2)+indC | 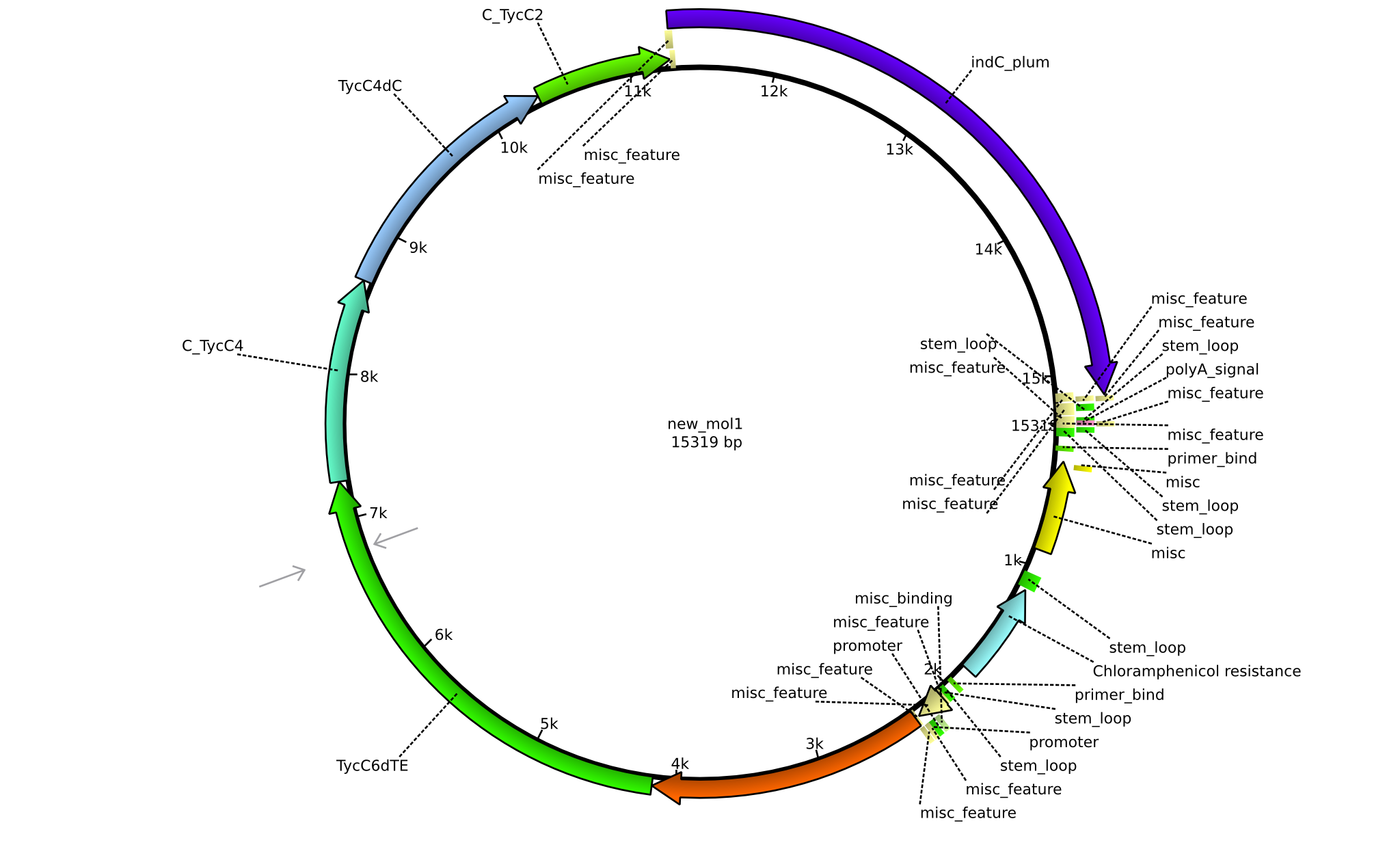 | pPW10 |
| pPW11 | 2013-09-07 | NRPS for Phe-Orn-Leu-Indigoidine | pSB1C3+TycAdCom+TycC5-TycC6dTE+C(TycC2)+indC | 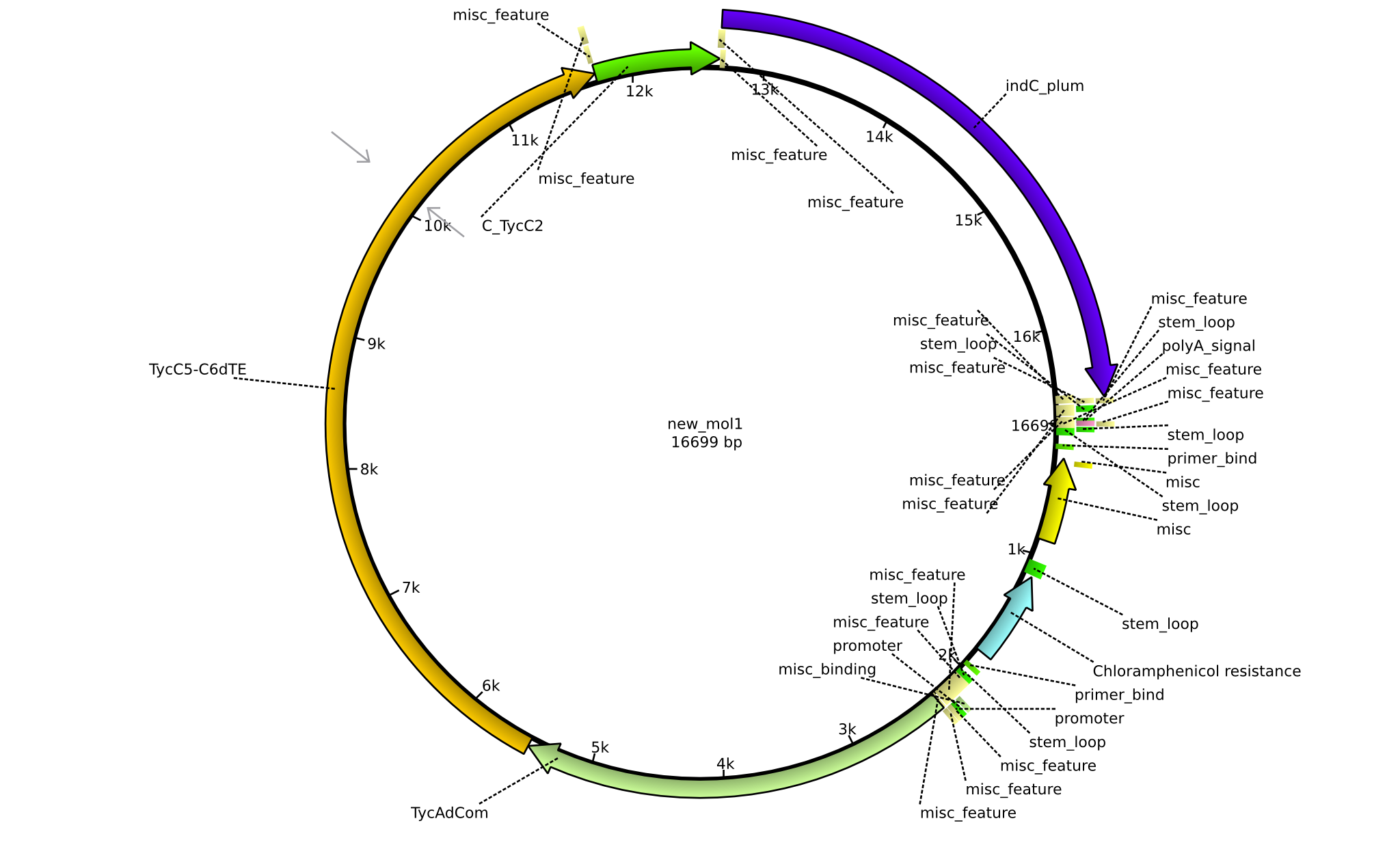 | pPW11 |
| pPW12 | 2013-09-07 | NRPS for Phe-Orn-Leu-Val-Indigoidine | pSB1C3+TycAdCom+TycC5-TycC6dTE+C(TycC4)+TycC4dC+C(TycC2)+indC | 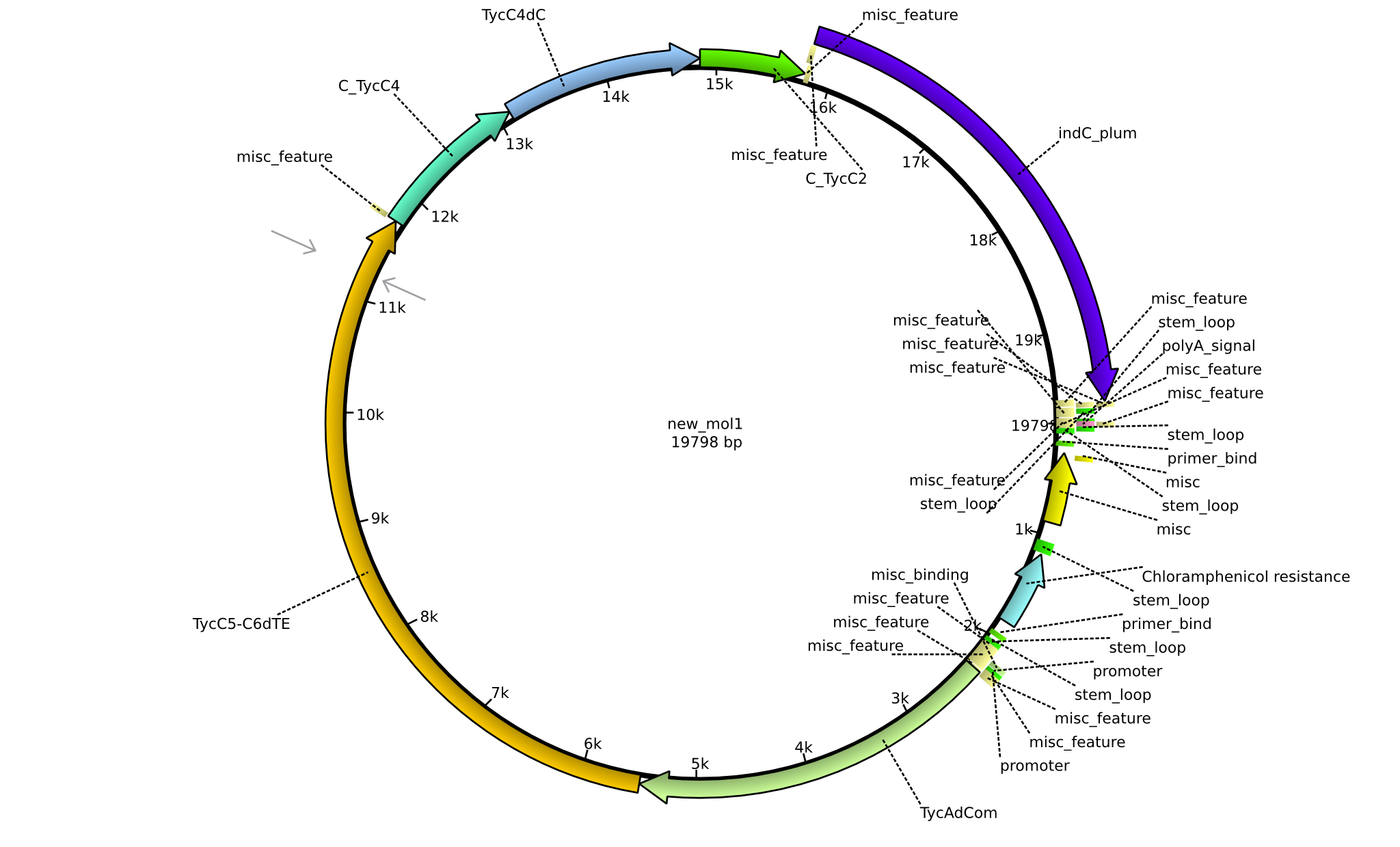 | pPW12 |
| pJS01 | 2013-09-18 | Helper plasmid for the tagging of NRPs with Indigoidine | pSB1C3+ccdB+C(TycC2)+indC | 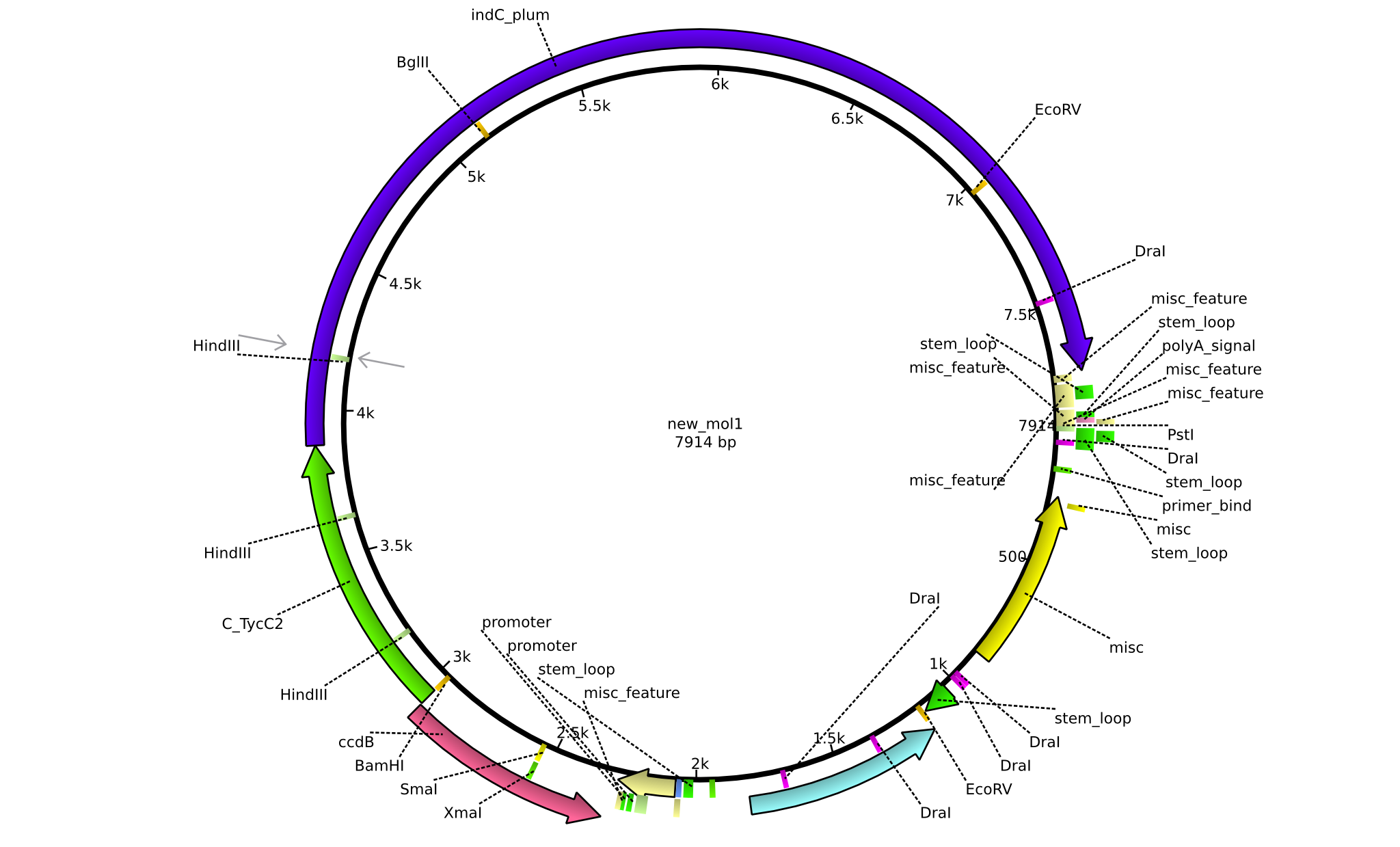 | pJS01 |
Linker variation
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
| pAR01 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with double short linkers | pSB1C3-TycC4dCom_s+TycC2_s+indC | 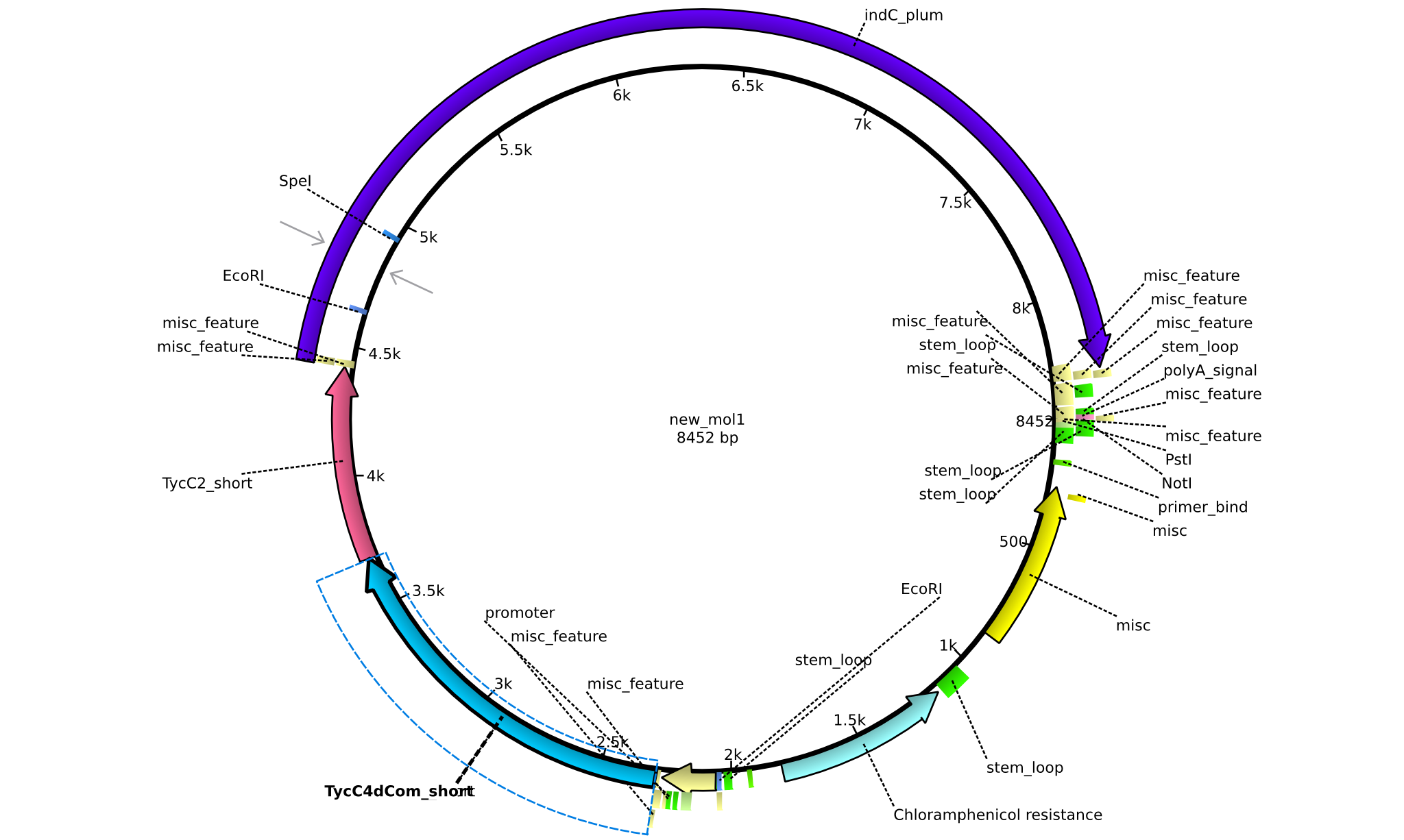 | pAR01 |
| pAR02 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C4 short linker | pSB1C3-TycC4dCom_s+TycC2_m+indC | 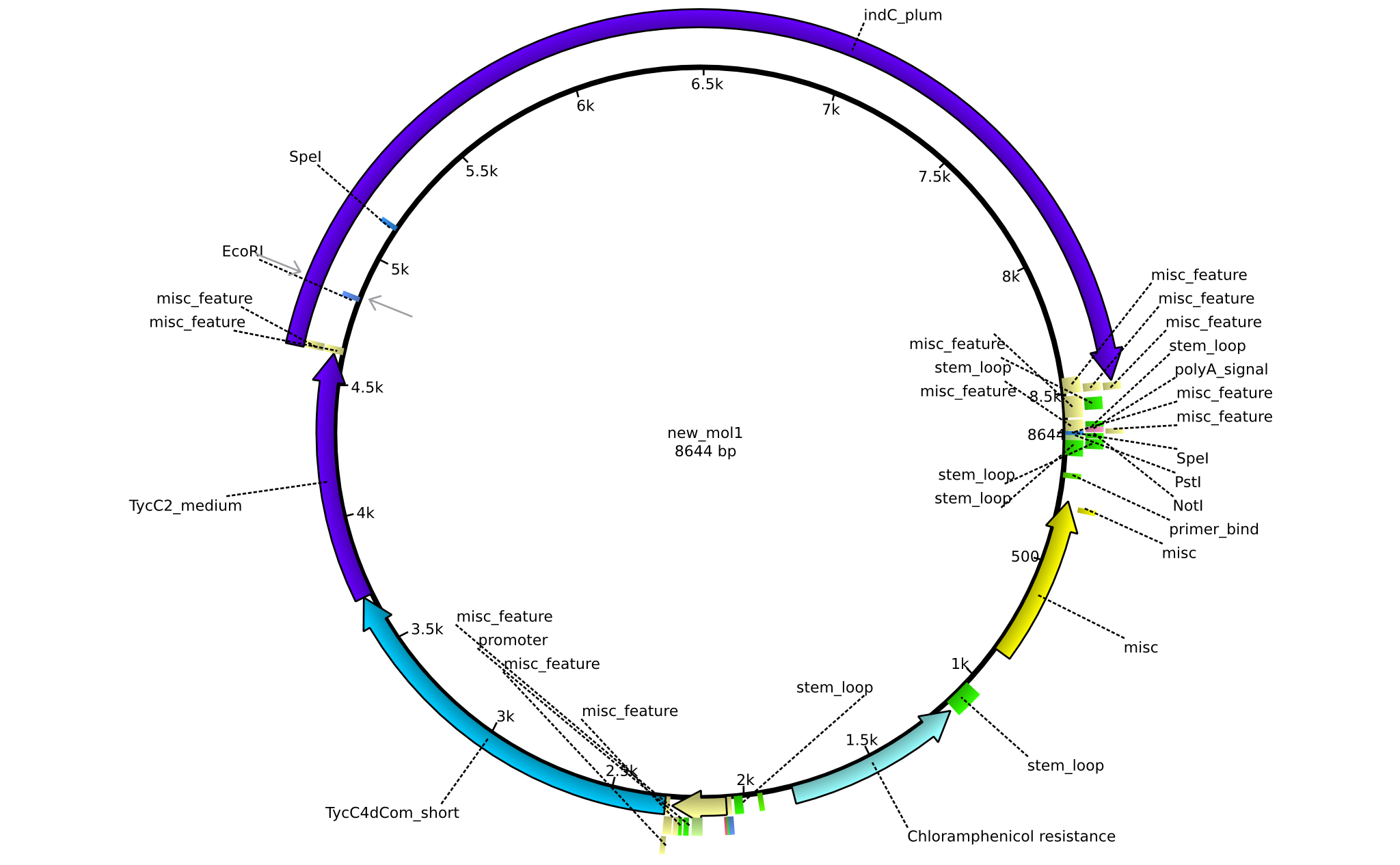 | pAR02 |
| pAR03 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C2 short linker | pSB1C3-TycC4dCom_m+TycC2_s+indC | 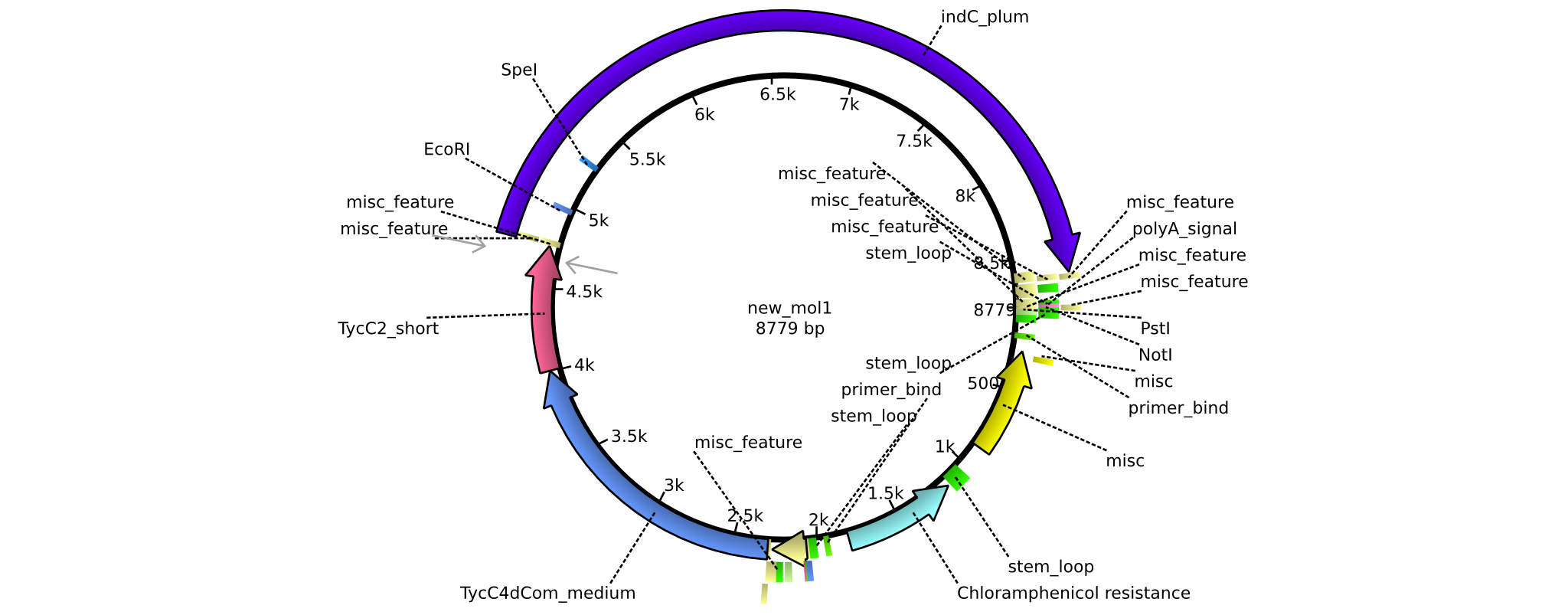 | pAR03 |
| pAR04 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with medium linkers | pSB1C3-TycC4dCom+TycC2+indC | 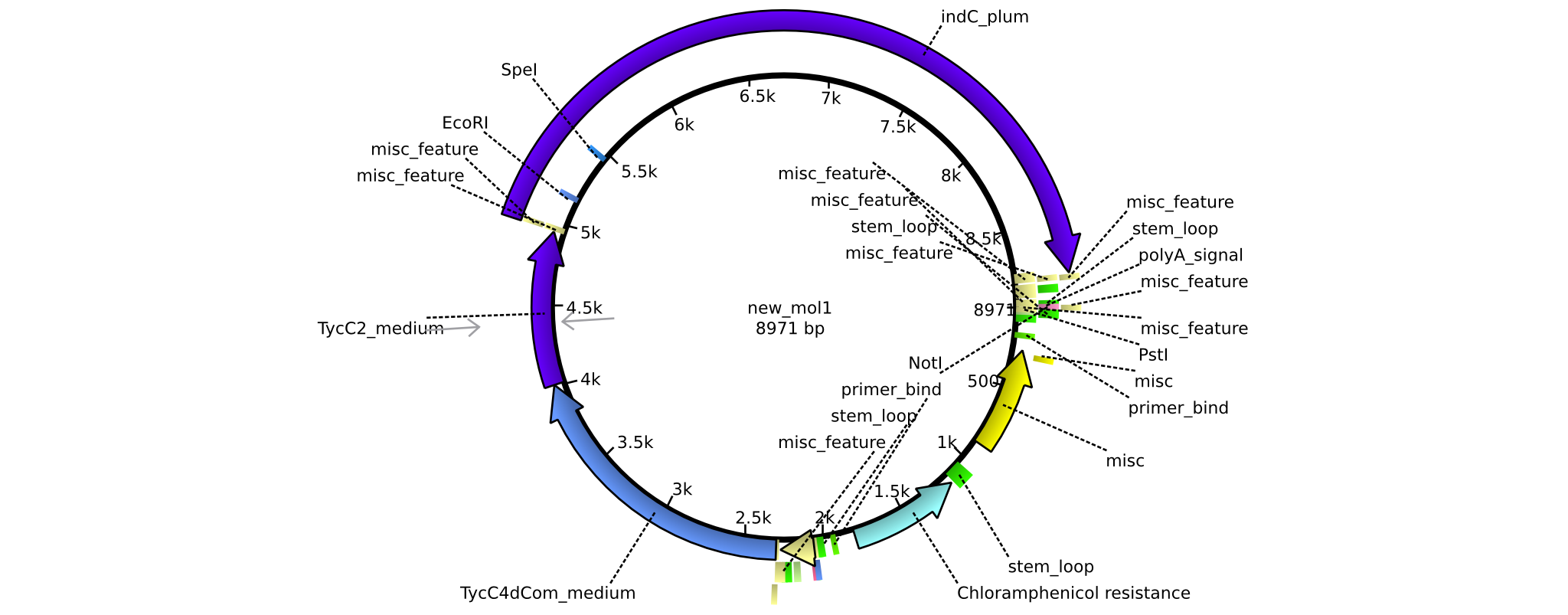 | pAR04 |
| pAR05 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C2 long linker | pSB1C3-TycC4dCom_m+TycC2_l+indC | 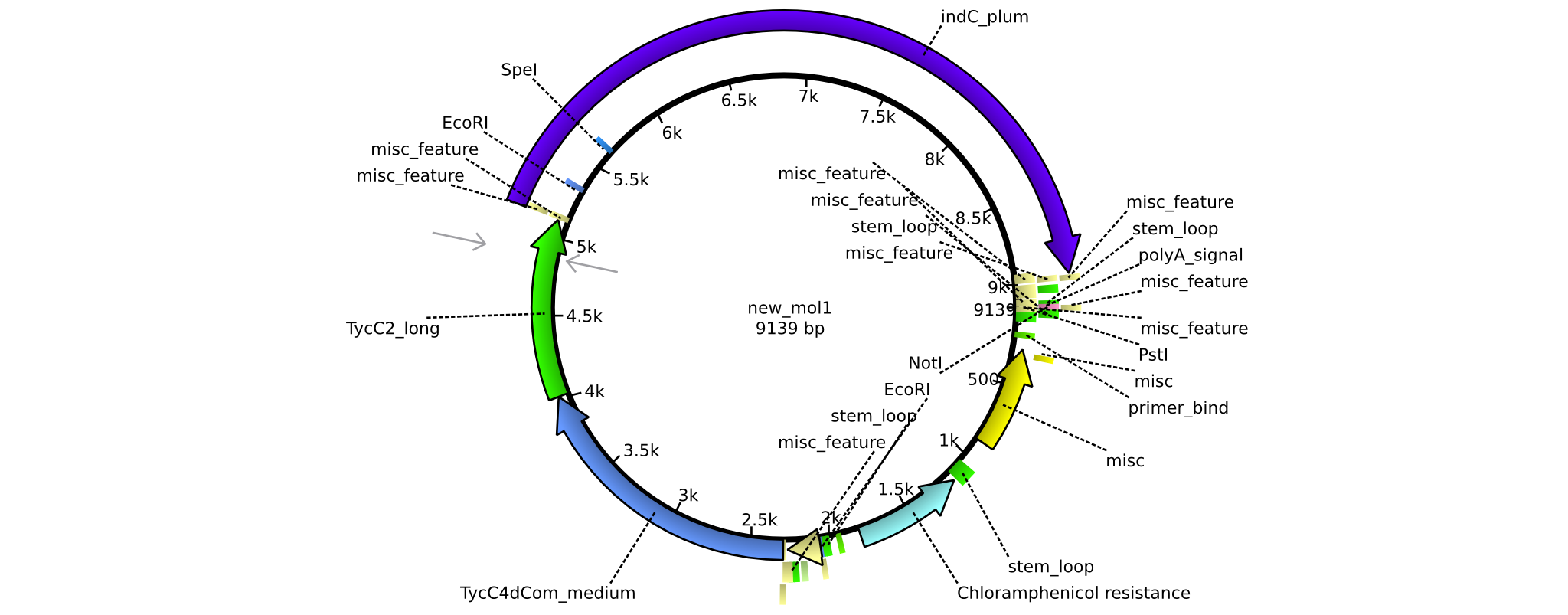 | pAR05 |
| pAR06 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C4 long linker | pSB1C3-TycC4dCom_l+TycC2_m+indC | 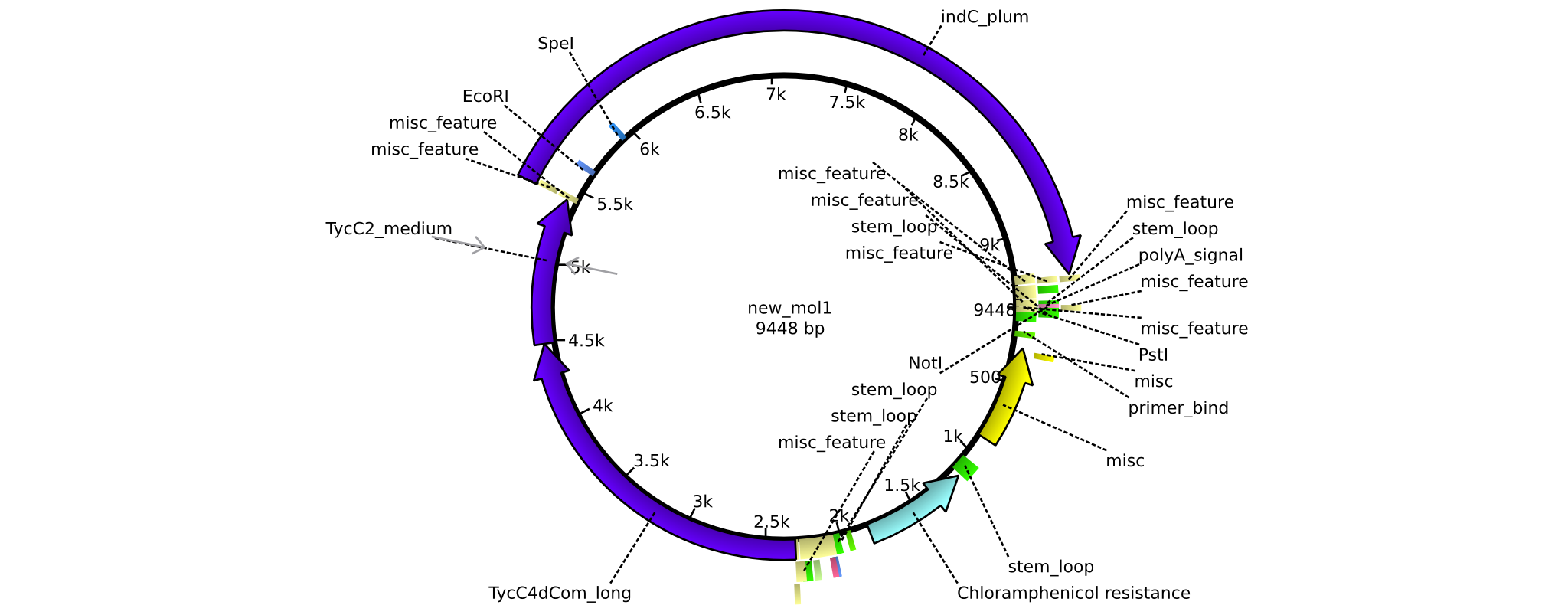 | pAR06 |
| pAR07 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with long linkers | pSB1C3-TycC4dCom_l+TycC2_l+indC | 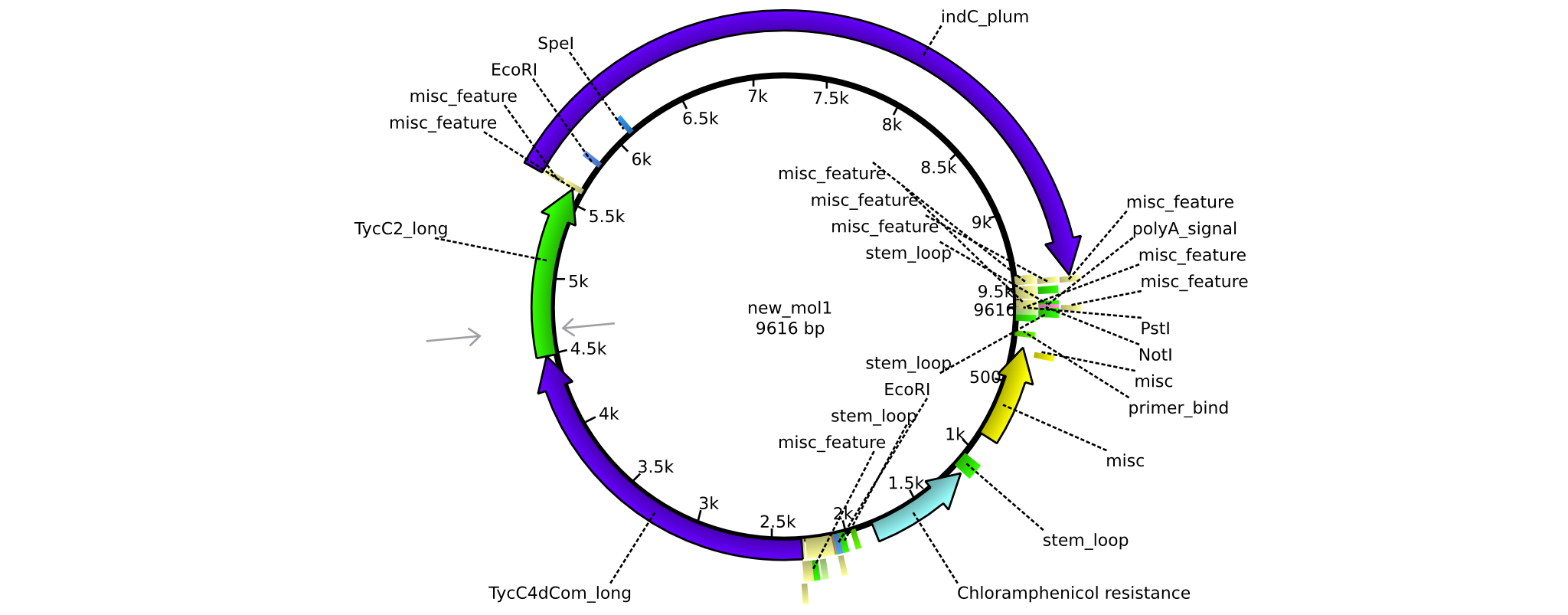 | pAR07 |
| pAR08 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C4 short and C2 long linker | pSB1C3-TycC4dCom_s+TycC2_l+indC | 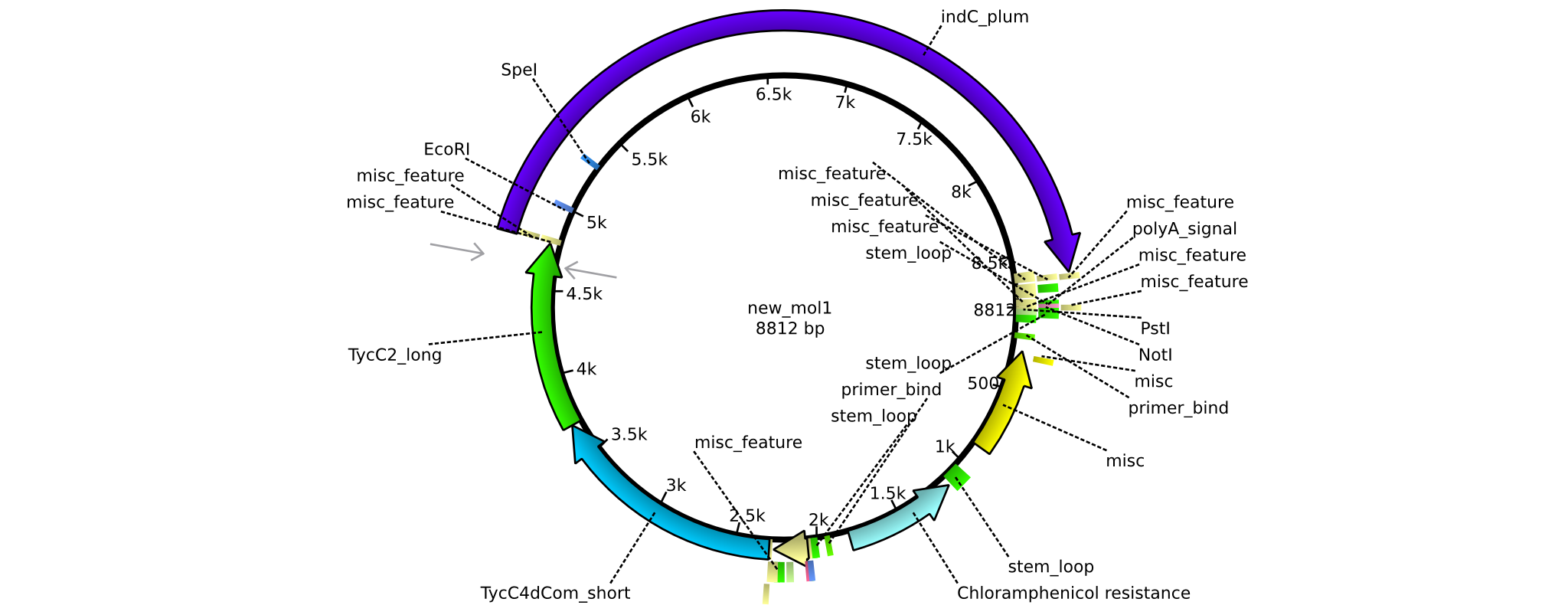 | pAR08 |
| pAR09 | 2013-16-09 | NRPS for Valin-Asparagine-Indigoidine-Expression with C4 long and C2 short linker | pSB1C3-TycC4dCom_l+TycC2_s+indC | 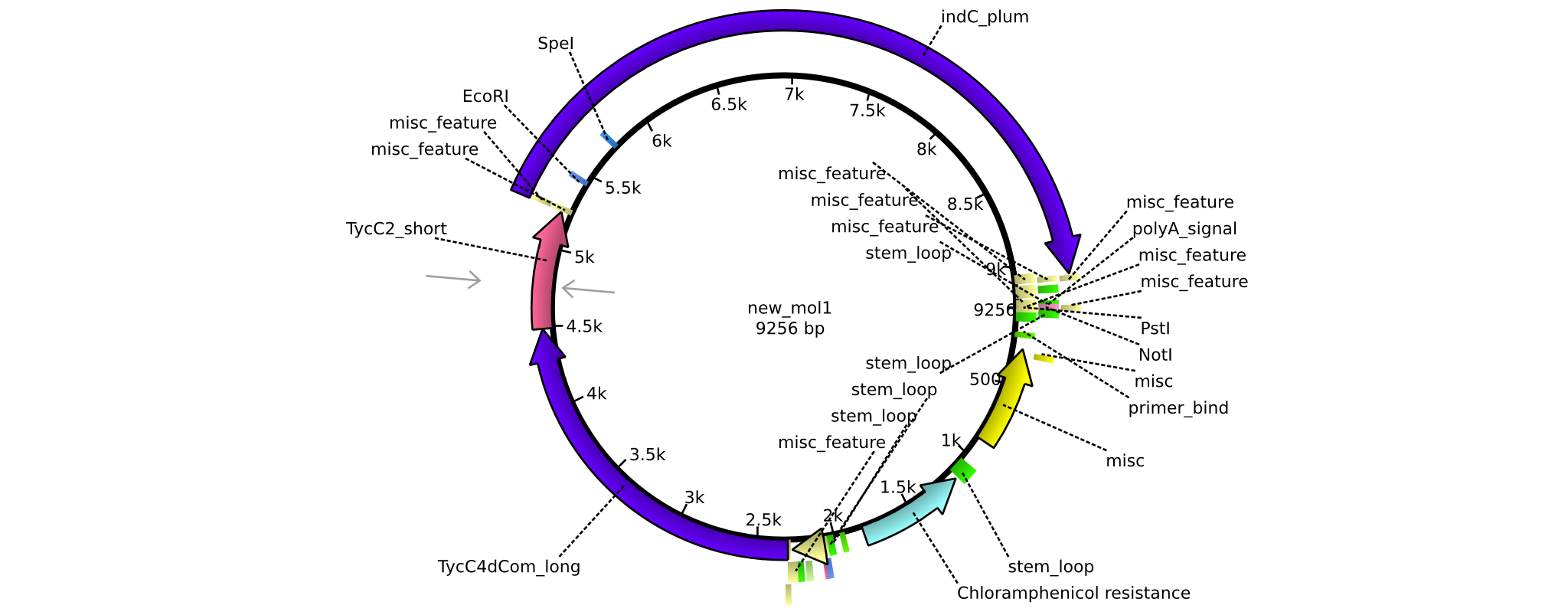 | pAR09 |
Dinner plasmids
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
pIK6 | 2013-08-27 | pIK6: special Ilia plasmid. This plasmid does not fullfill iGEM's RFC10 but is nevertheless special. It is for expression of tasty Ham driven by a strong lacCheese inducible promoter, a RBSalami for increased fleshiness, which can only terminated by a Champignon stop codon. The pIZa backbone with Pepper resistance and high copy Onion ori will improve the users taste stimulation. Bon Appétit! | pIZa-lacCheese P-RBSalami-ham CDS-Champ Term-Pepper res-Onion Ori |  | pIK6 |
Domain Shuffling and PPTases
| Name | Date | Brief Description | Genotype | Plasmid Map | GenBank-File |
|---|---|---|---|---|---|
pMM64 | 2013-06-01 | bpsA(fus) | pETDuet-1-T7lac-bpsA(fus) |  | pMM64 |
pMM65 | 2013-06-01 | svp(fus) | pETDuet-1-T7lac-svp(fus) |  | pMM65 |
pKH4 | 2013-09-01 | pRB3/pMM64 derived plasmid without sfp | pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-nonsense |  | pKH4 |
pKH6 | 2013-09-05 | BBa CDS sfp only for part submission | pSB1C3-sfp | 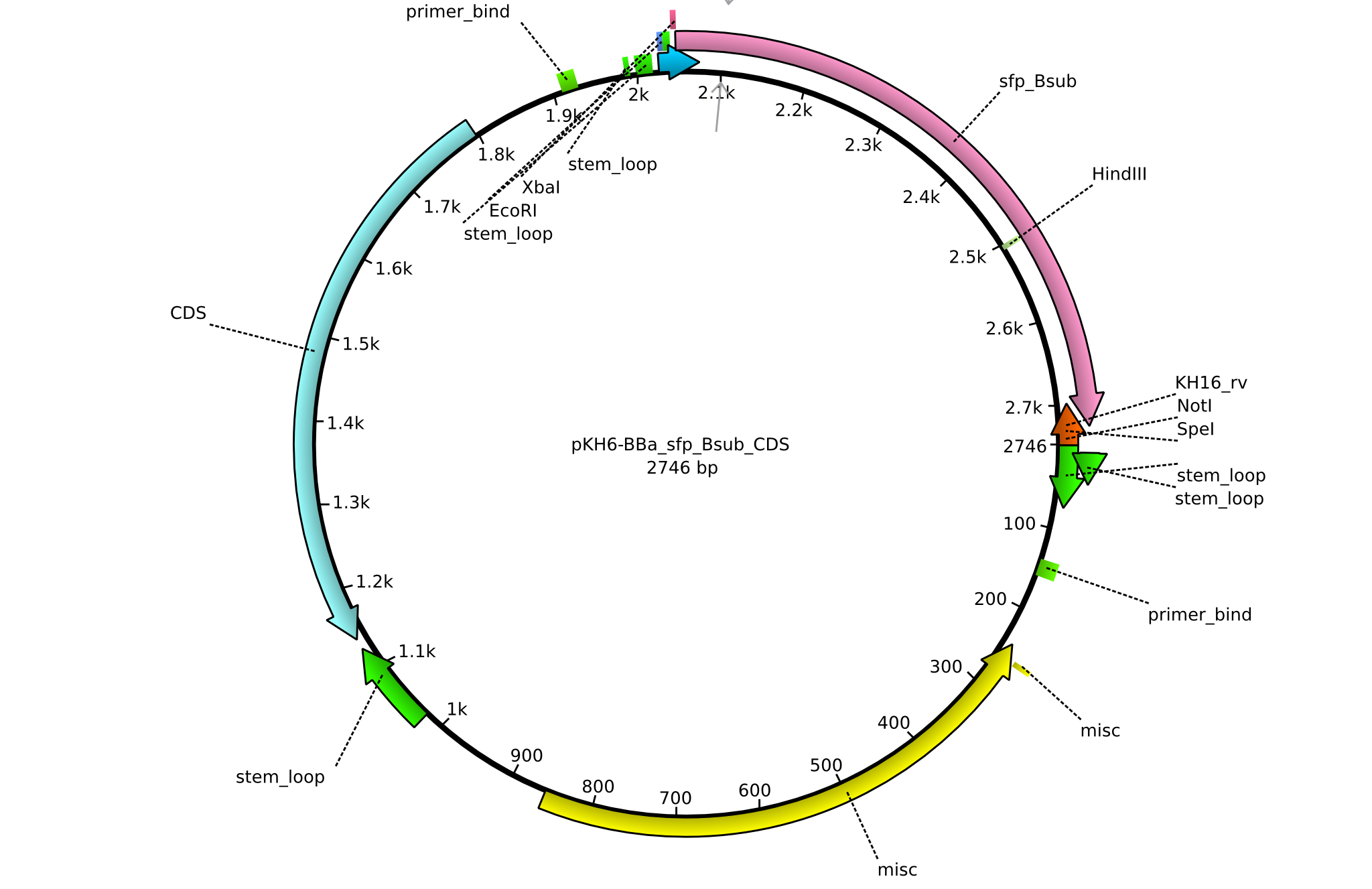 | pKH6 |
pKH7 | 2013-09-05 | BBa CDS svp only for part submission | pSB1C3-svp | 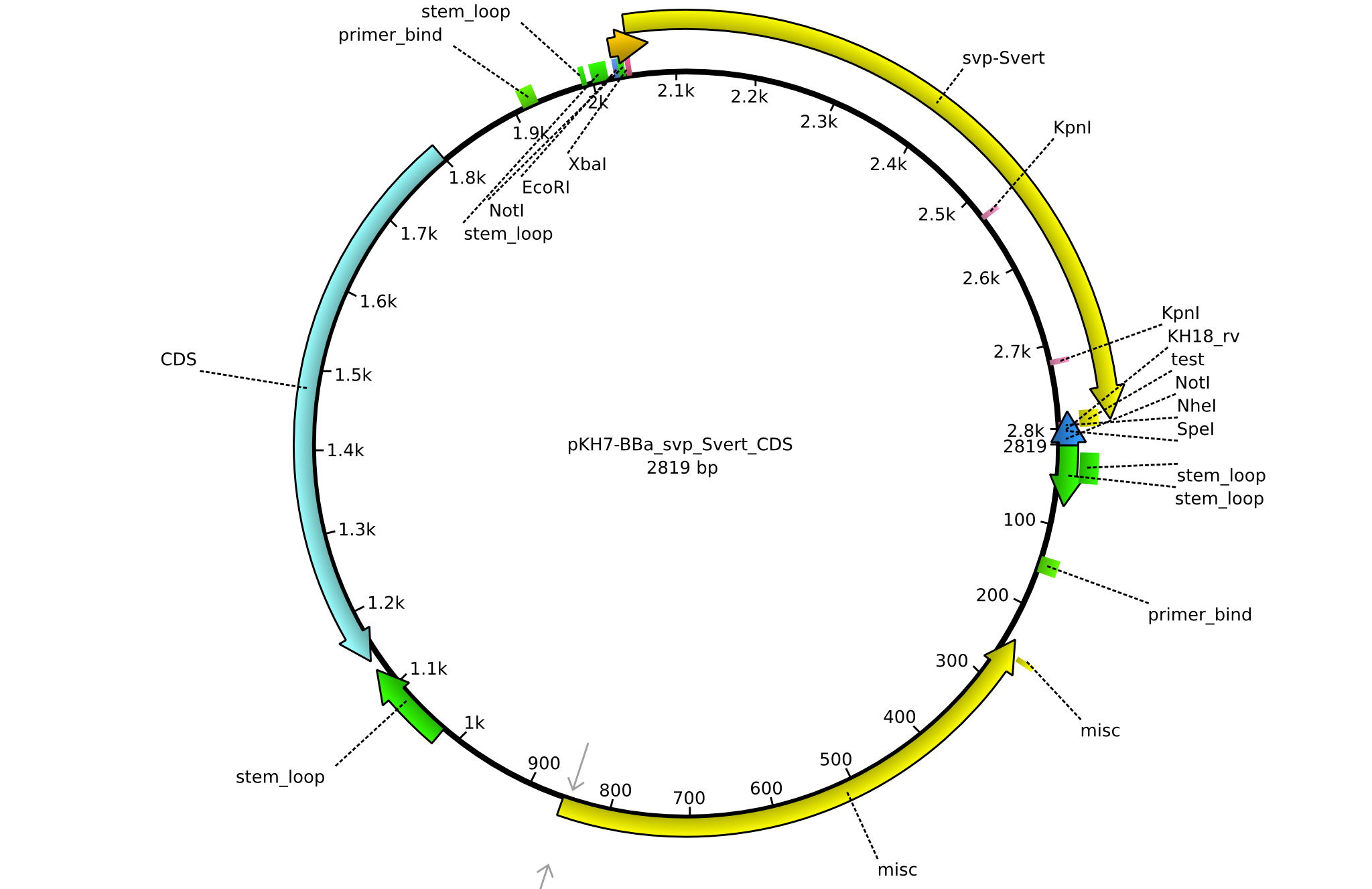 | pKH7 |
pKH8 | 2013-09-05 | BBa CDS entD only for part submission | pSB1C3-entD | 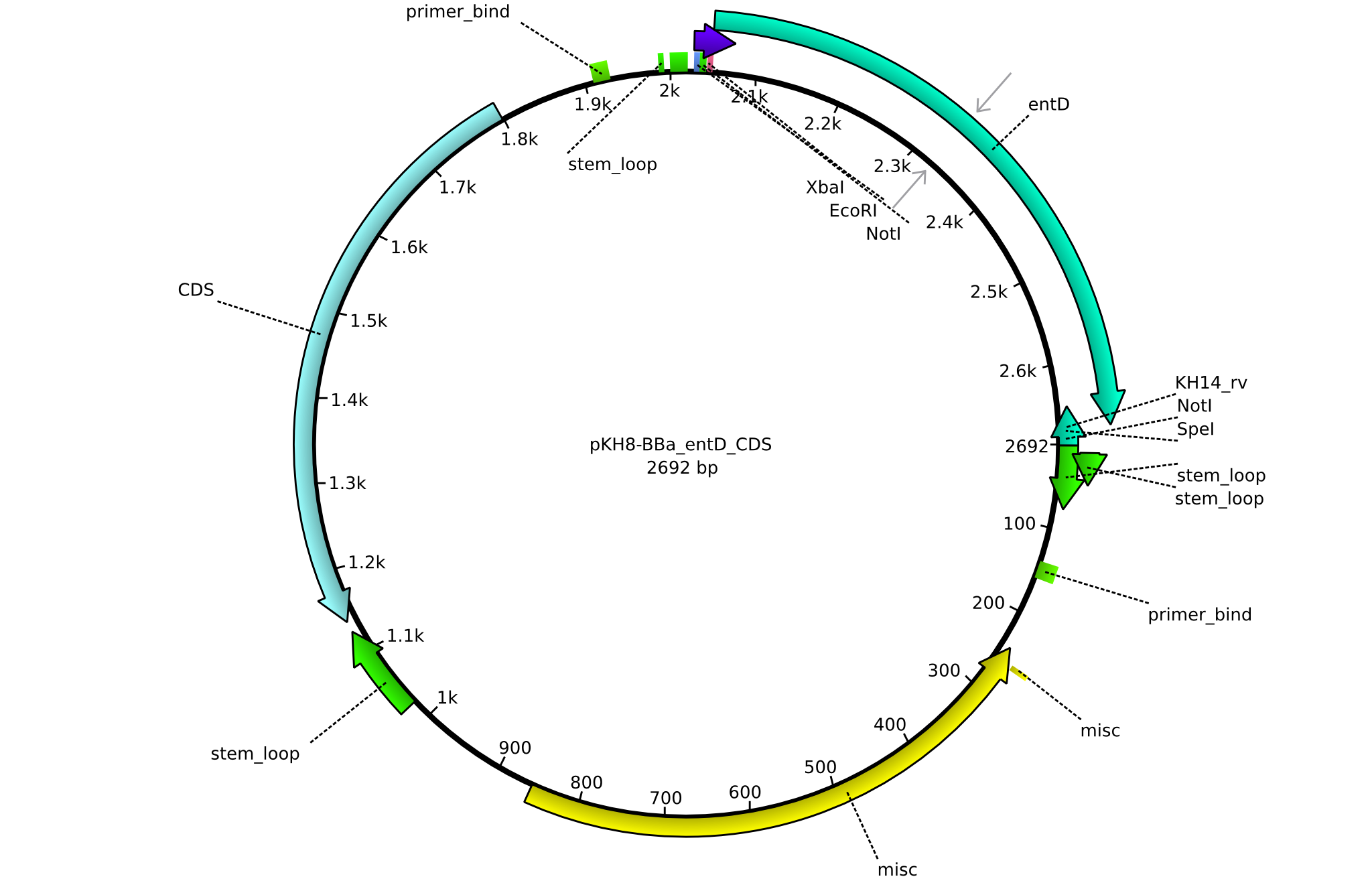 | pKH8 |
pKH9 | 2013-09-05 | BBa CDS delC only for part submission | pSB1C3-delC | 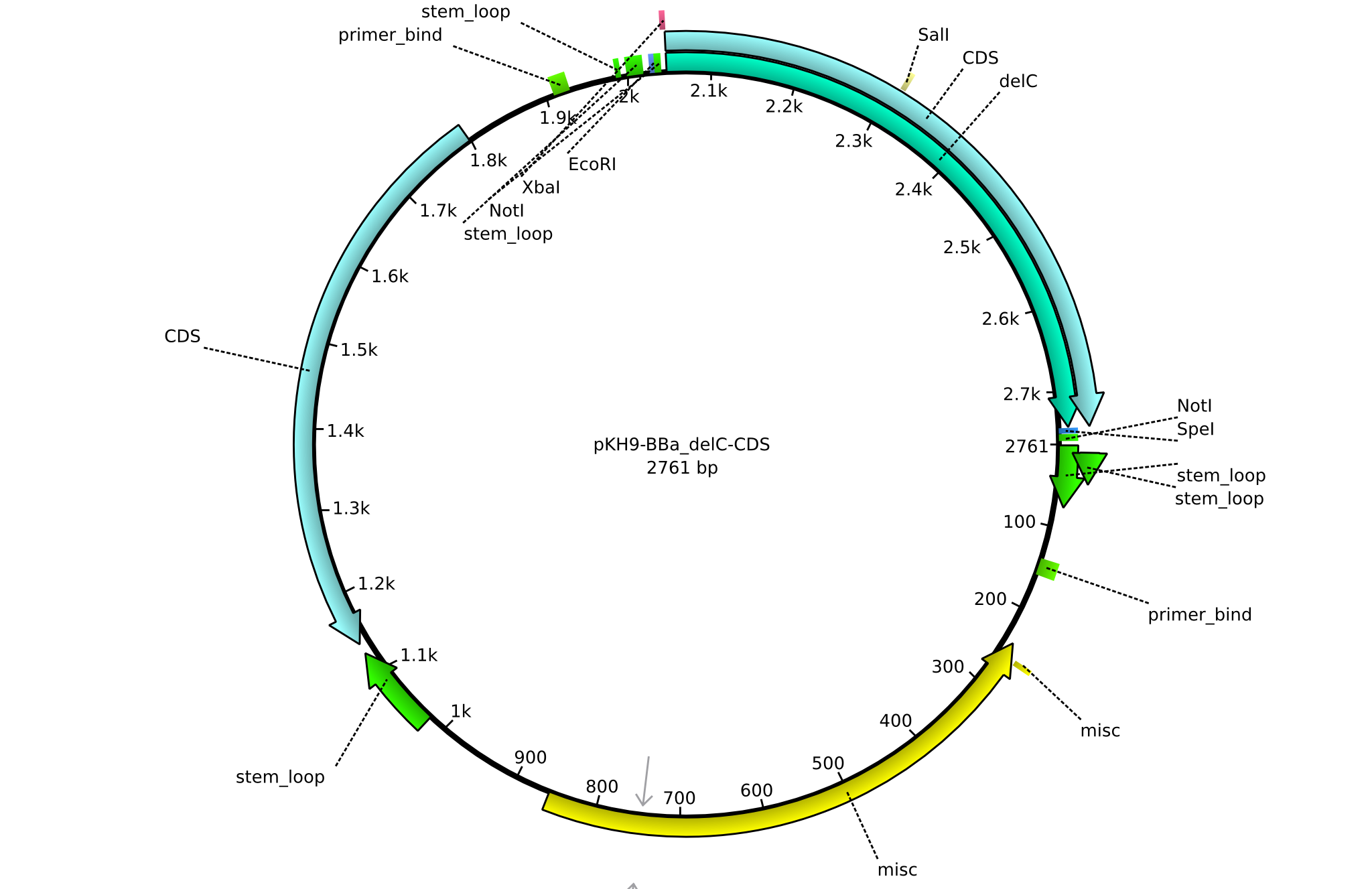 | pKH9 |
pRB1 | 2013-07-01 | bpsA svp(pMM65) | pSB1C3-lacP-BBa_B0034-bpsA-BBa_B0029-svp(pMM65) | N.A | N.A |
pRB2 | 2013-07-08 | bpsA svp | HindIII-pSB1C3-lacP-BBa_B0034-bpsA-BBa_B0029-HindIII-svp | N.A | N.A |
pRB3 | 2013-07-15 | indC sfp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-BamHI-sfp | 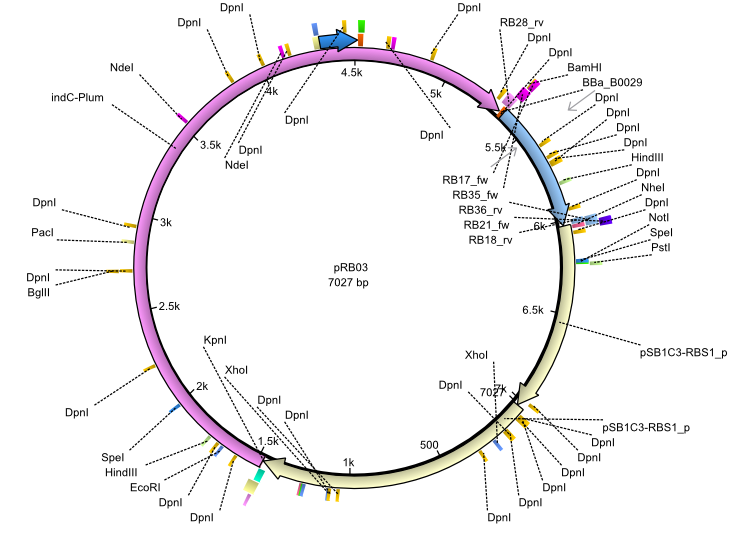 | pRB03 |
pRB4 | 2013-07-15 | indC svp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-BamHI-svp | 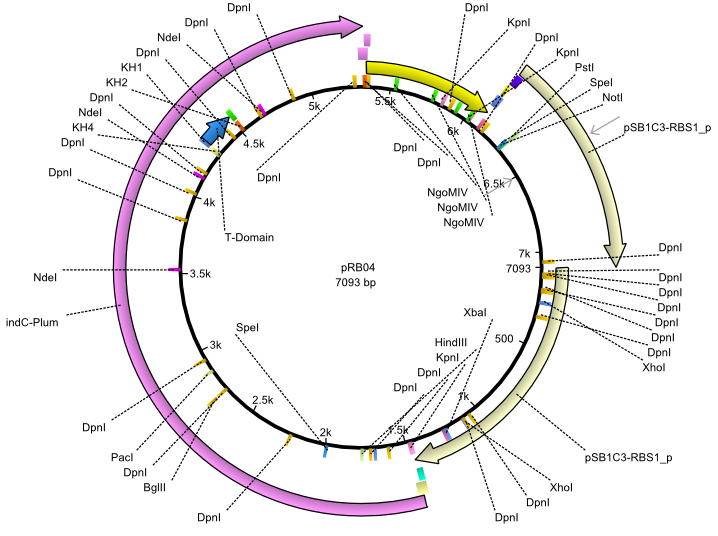 | pRB04 |
pRB5 | 2013-07-15 | indC svpF | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-BamHI-svp(pMM65) | 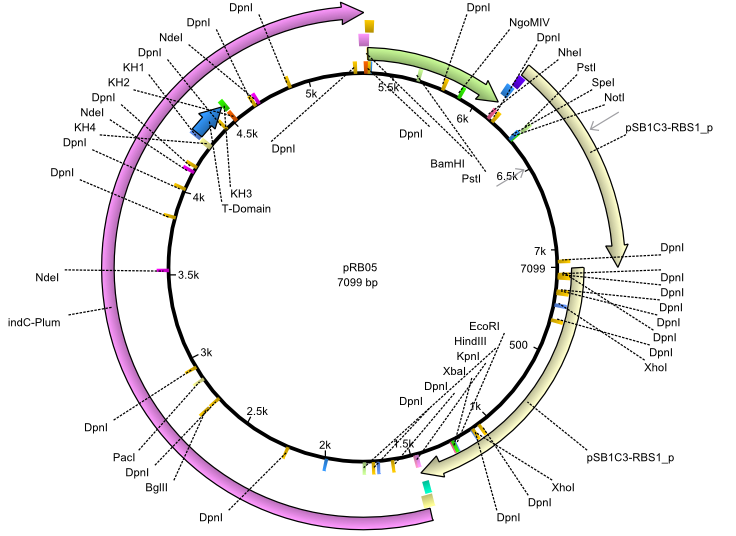 | pRB05 |
pRB6 | 2013-07-15 | indC entD | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-BBa_B0029-BamHI-entD | 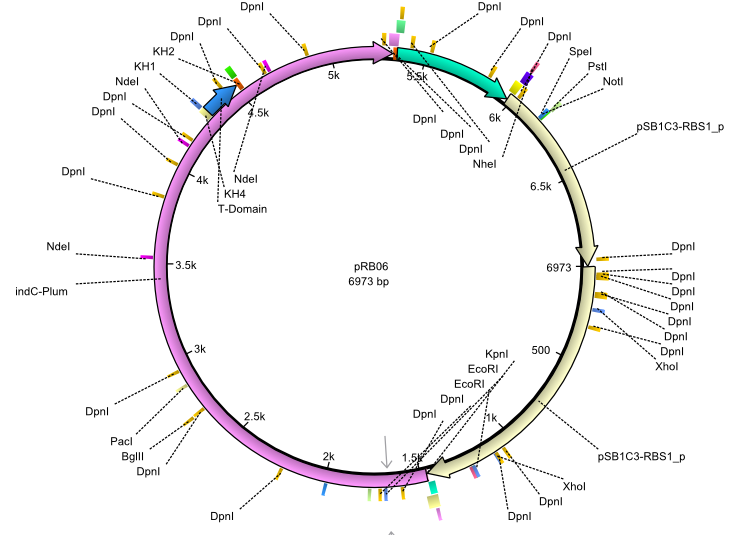 | pRB06 |
pRB7 | 2013-07-15 | bpsA(pMM64) sfp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-bpsA(pMM64)-BBa_B0029-BamHI-sfp | 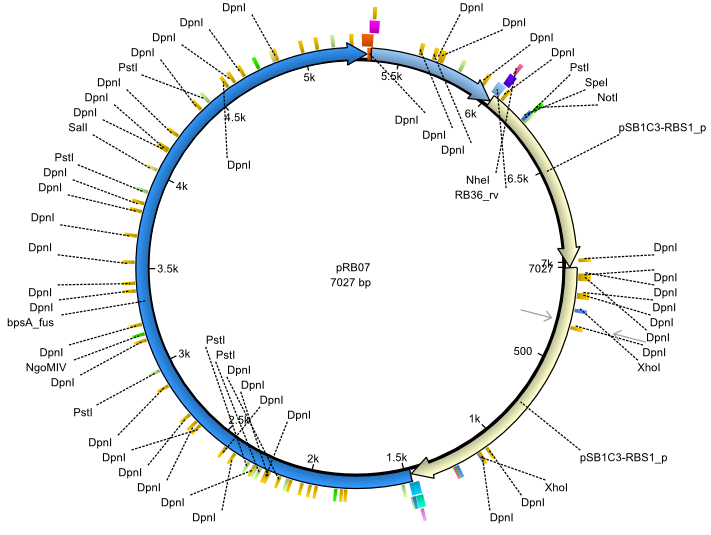 | pRB07 |
pRB8 | 2013-07-15 | bpsA(pMM64) svpF | NheI-pSB1C3-lacP-BBa_B0034-KpnI-bpsA(pMM64)-BBa_B0029-BamHI-svp(pMM65) | 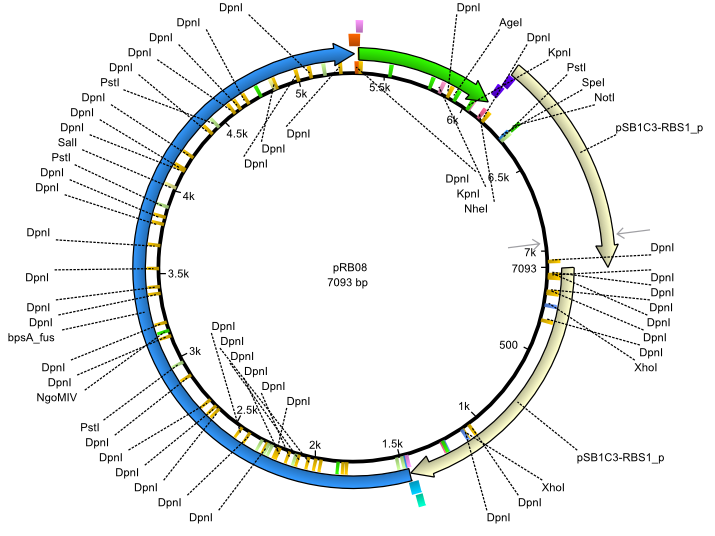 | pRB08 |
pRB9 | 2013-07-15 | bpsA(pMM64) svp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-bpsA(pMM64)-BBa_B0029-BamHI-svp | 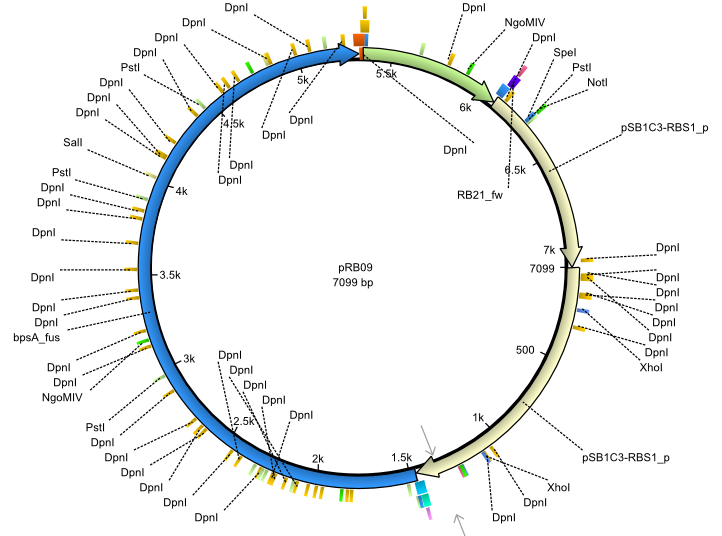 | pRB09 |
pRB10 | 2013-07-15 | bpsA(pMM64) entD | NheI-pSB1C3-lacP-BBa_B0034-KpnI-bpsA(pMM64)-BBa_B0029-BamHI-entD | 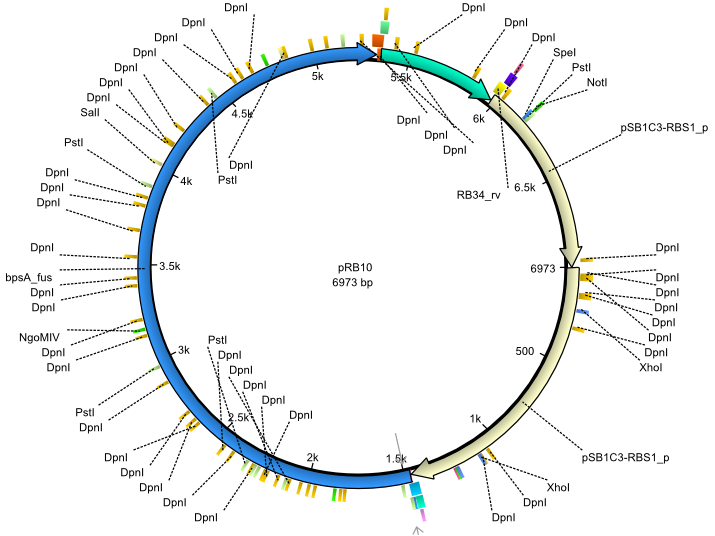 | pRB10 |
pRB11 | 2013-07-29 | pKH1-der bpsA(pMM64)-ccdb svpF | pSB1C3-lacP-BBa_B0034-bpsA(pMM64)(ccdb)-BBa_B0029-svp(pMM65) | N.A | N.A |
pRB12 | 2013-07-29 | pKH2-der bpsA(pMM64)-ccdb svpF | pSB1C3-lacP-BBa_B0034-bpsA(pMM64)(ccdb)-BBa_B0029-svp(pMM65) | N.A | N.A |
pRB13 | 2013-07-29 | pRB3-der indC-ccdb sfp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC(ccdb)-BBa_B0029-BamHI-sfp | N.A | N.A |
pRB14 | 2013-08-12 | pRB3-der indC-ccdB sfp | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC(ccdB)-BBa_B0029-BamHI-sfp | N.A | N.A |
pRB15 | 2013-08-12 | pSB3K3 sfp | pSB3K3-lacP-BBa_B0029-BamHI-sfp-NheI | 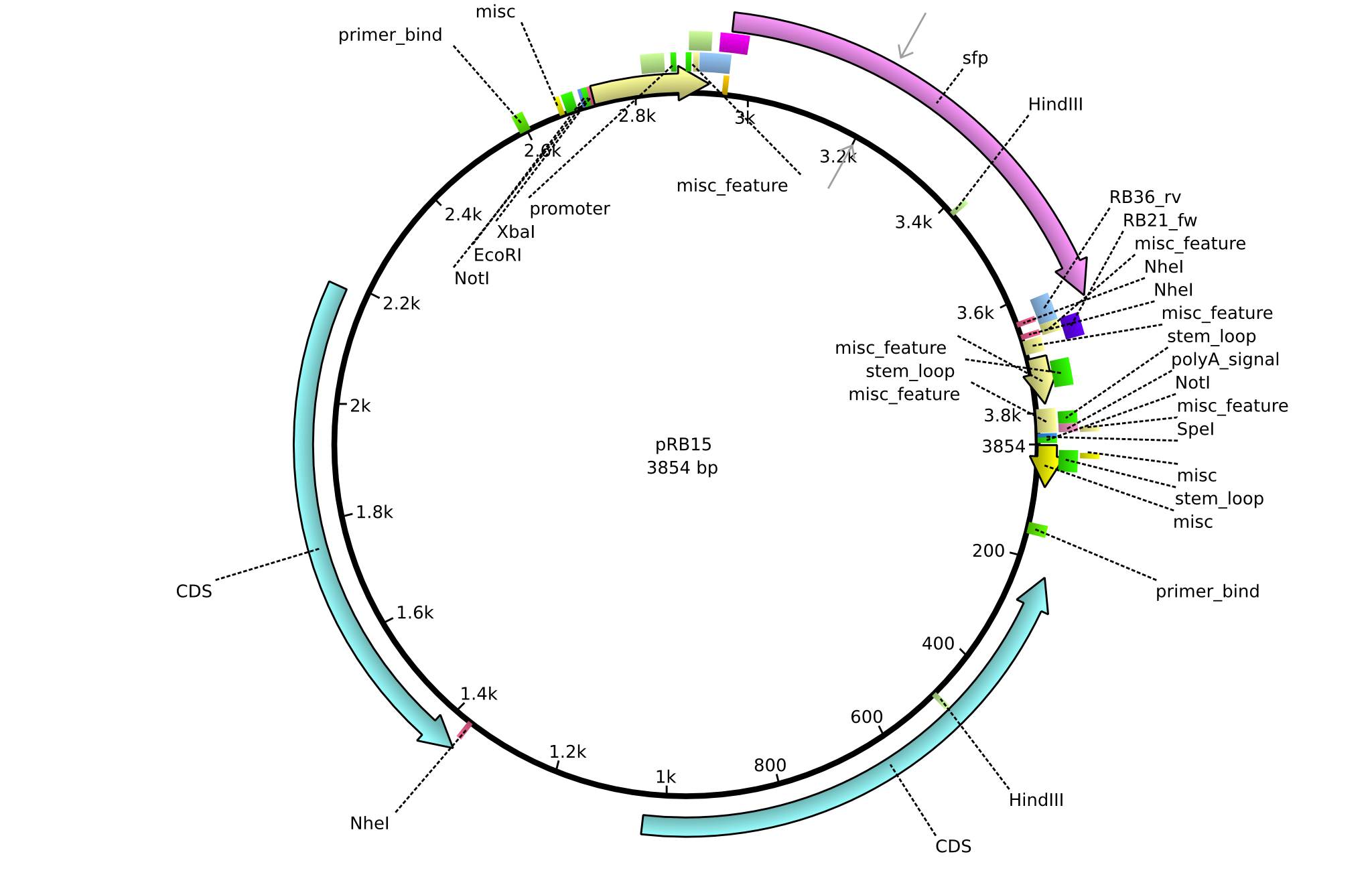 | pRB15 |
pRB16 | 2013-08-12 | pSB3K3 svp | pSB3K3-lacP-BBa_B0029-BamHI-svp-NheI | 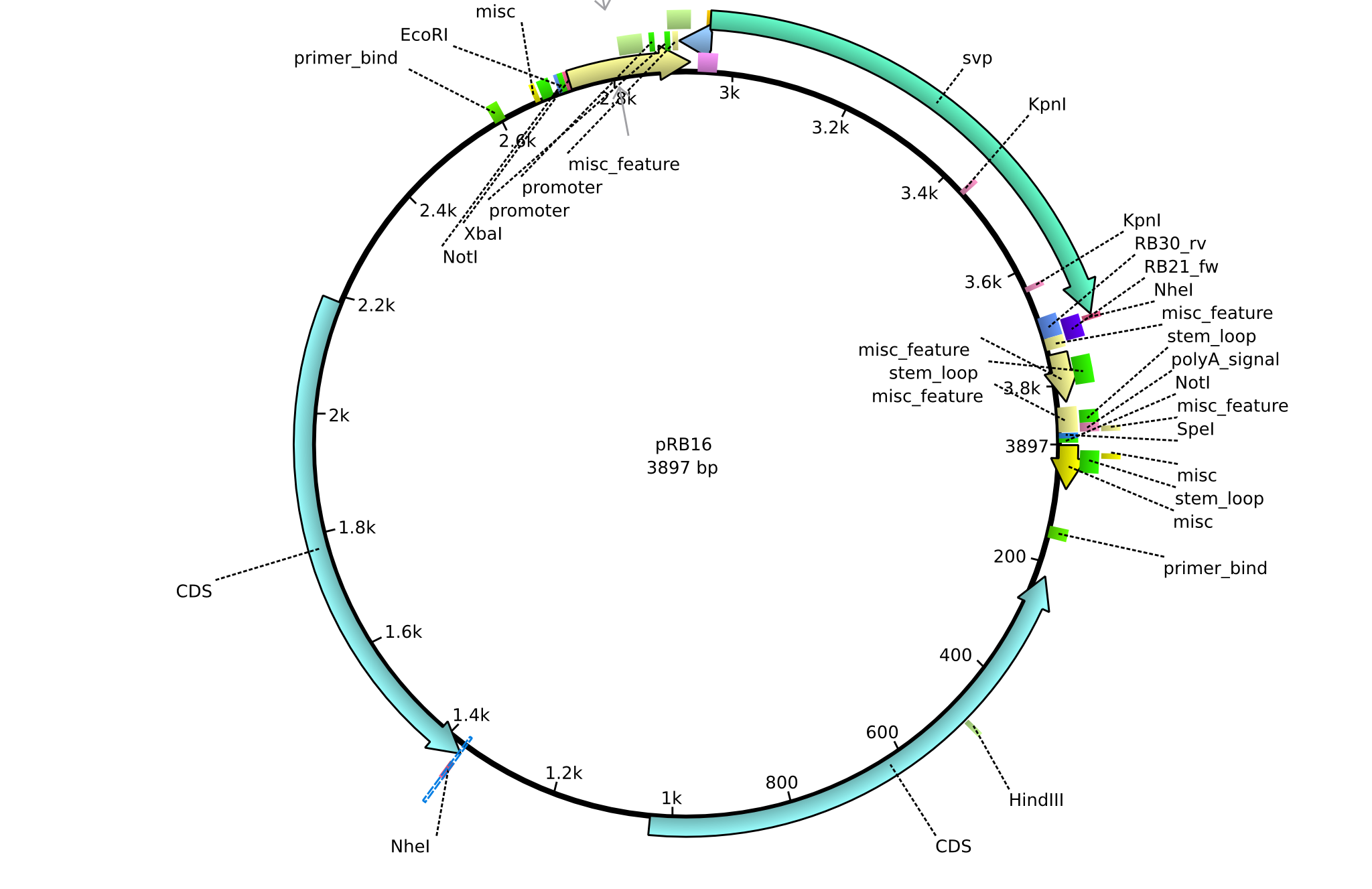 | pRB16 |
pRB17 | 2013-08-12 | pSB3K3 entD | pSB3K3-lacP-BBa_B0029-BamHI-entD-NheI | 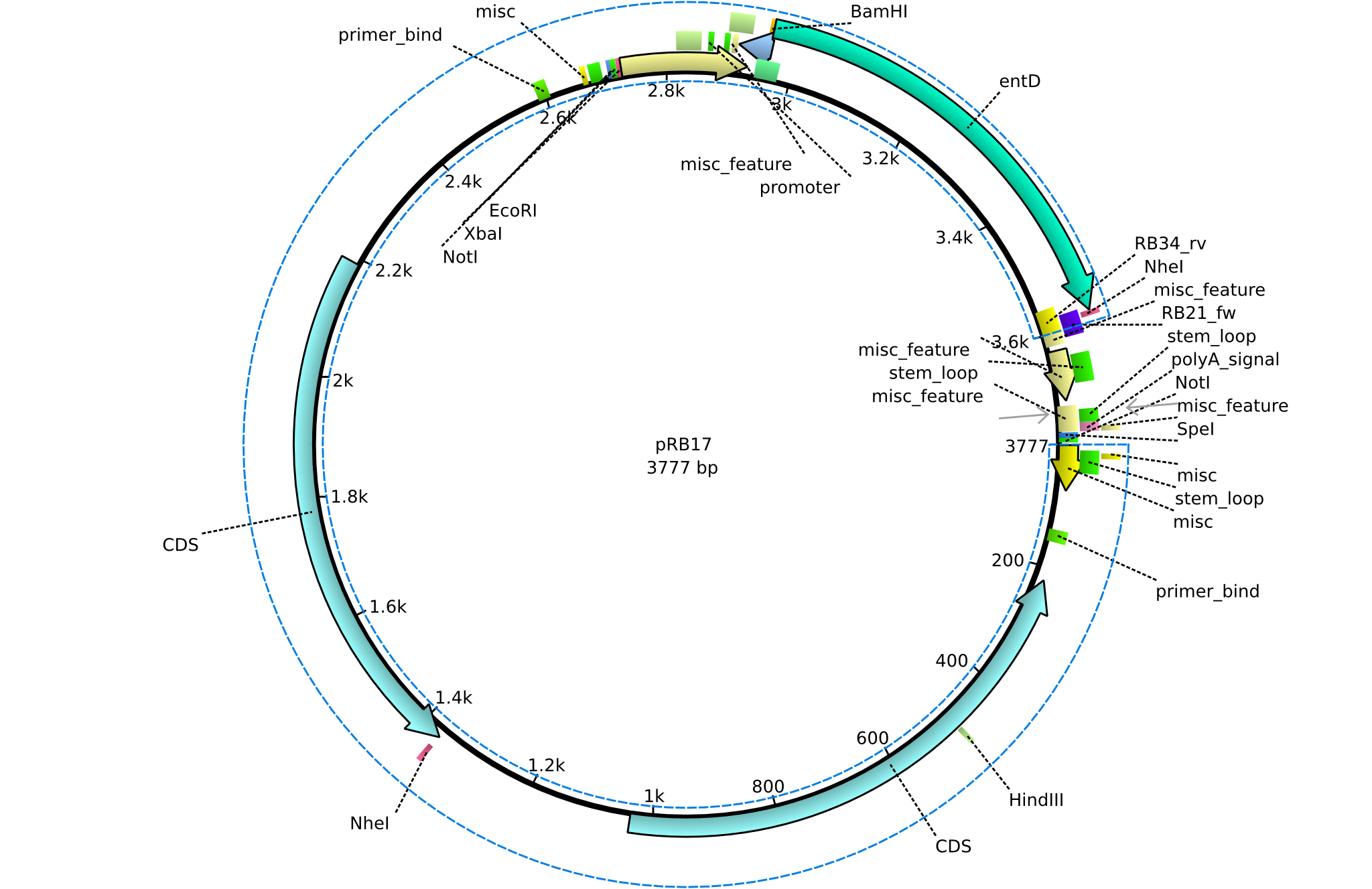 | pRB17 |
pRB18 | 2013-08-12 | pSB3K3 delC | pSB3K3-lacP-BBa_B0029-BamHI-delC-NheI | 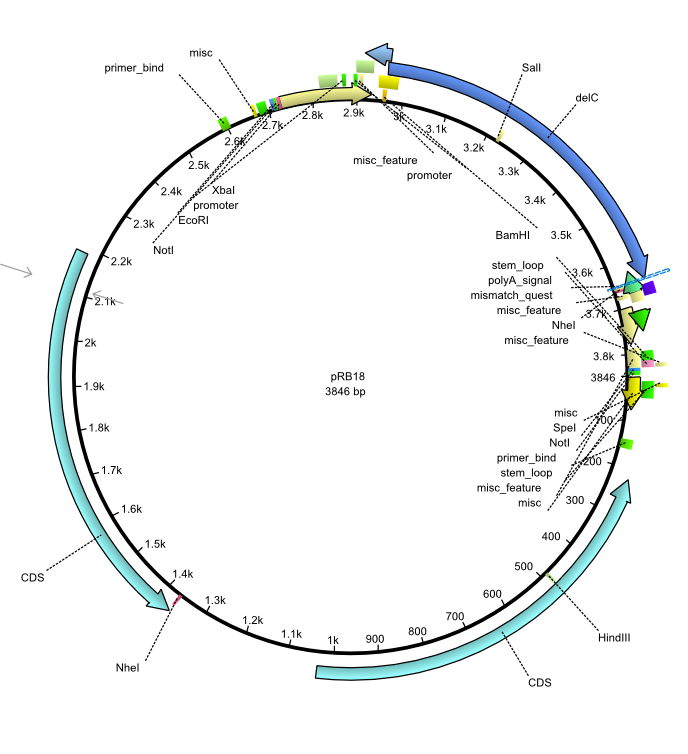 | pRB18 |
pRB19 | 2013-08-19 | pRB14-der indC-ccdB | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC(ccdB) | N.A | N.A |
pRB20 | 2013-08-19 | pRB19-der indC-HD-ccdB | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-HD(ccdB) | N.A | N.A |
pRB21 | 2013-08-19 | pSB1C3 indC | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC |  | pRB21 |
pRB22 | 2013-08-19 | pRB21-der indC-HD | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC-HD |  | pRB22 |
pRB23 | 2013-08-19 | pRB22-der indC-HD | NheI-pSB1C3-lacP-BBa_B0034-KpnI-indC*dT(ccdB) | N.A | N.A |
Used T and TE Domains
| ID | Name (derived from) | Description |
|---|---|---|
-T1 | indC | native T-Domain of indC from P. luminescens |
-T2 | bpsA | native T-Domain of bpsA from S. lavendulae |
-T3 | entF | native T-Domain of entF from E. coli |
-T4 | tycA1 | native T-domain of tycA1 from Brevibacillus parabrevis |
-T5 | tycC6 | native T-domain of tycC6 from B. parabrevis |
-T6 | delH4 | native T-domain of delH4 from Delftia acidovorans |
-T7 | delH5 | native T-domain of delH5 from D. acidovorans |
-T8 | plu2642 | native T-domain of plu2642 from Photorhabdus luminescens |
-T9 | plu2670 | native T-domain of plu2670 from P. luminescens |
-T10 | synT1 | synthetic T-Domain from indC-BLAST |
-T11 | synT2 | synthetic T-Domain from indC_T-Plum-BLAST |
-T12 | synT3 | synthetic T-Domain from indC_T-BLAST |
-T13 | synT4 | synthetic T-Domain from project_5 BLAST |
-T14 | synT5 | synthetic T-Domain random 1 (indigoidine synthetases) |
-T15 | synT6 | synthetic T-Domain random 2 (project_5) |
-TTE1 | bpsA | native TTE-Domain of bpsA from S. lavendulae |
-TTE2 | entF | native TTE-Domain of entF from E. coli |
-TTE3 | tycC6 | native TTE-domain of tycC6 from B. parabrevis |
-TTE4 | delH5 | native TTE-domain of delH5 from D. acidovorans |
Instruments
| Instrument | Type | Manufacturer |
|---|---|---|
| Table Centrifuge | Microfuge® 18 Centrifuge | Beckman CoulterTM |
| Table Centrifuge | Microfuge® 22R Centrifuge | Beckman CoulterTM |
| Centrifuge | Allegra X-12R | Beckman CoulterTM |
| Shaker | VORTEXGENIE 2 | Scientific Industries, SITM |
| Heating plate (magnet stirrer) | MR Hei-Standard | Heidolph |
| Heatblock | QBD4 | Grant |
| Heatblock (shakeing function) | Thermomixer comfort | Eppendorf |
| PCR-Machine | MyCyclerTM thermo cycler | BioRad |
| PCR-Machine | T100 Therma Cycler | BioRad |
| UV-Chamber | Transluminator | Vilber Lourmat |
| Scale (fine) | PioneerTM PA114C | OHAUS |
| Scale | PioneerTM PA4101C | OHAUS |
| Fridge | KTP 1750 Premium | Liebherr |
| Freezer | GP 1366 Premium | Liebherr |
| Hood | Tischabzug | Wesemann® Laboreinrichtung |
| Draw-Off Pump | Vacuhand control | Vacubrand |
| Incubator | HT Multitron Version 2 | INFORS |
| Plate Reader | ||
| Computer | Sun Microsystems | |
| UV/VIS Spectrometer | Ultrospec 3300 pro | Amersham Biosciences |
| Photometer | NanoDrop® ND-1000 Spectrophotometer | peQLab Biotechnologie GmbH |
| Photometer | NanoVue | General Electric |
| Ice Machine | MF22 | SCOTSMAN® |
| Gel Electrophoresis Chamber | Mupid®-One | Advance |
| Cell Density Meter | Ultrospec10 | Amersham Biosciences |
| PH-Meter | PH-Meter 765 Calimatic | Knick |
| Incubator | Heraeus | Thermo Scientific |
| Lyophilizer | Gefriertrocknungsanlage ALPHA 1-2 LD Plus Best. Nr. 101521 | Christ |
| Ultra Sonification Stick | Sonoplus Gm 2070 (2002) | Bandelin |
| Vacuum Manifold | Qiavac 24 Plus | QIAGEN |
| Gas Cartridge Ventil CV470 | Burner | 13883 |
| NuPAGE(R) Novex 3-8% Tris-Acetate Gel 1 | SDS-Gel | Life Technologies GmbH |
Mass Spectrometry and Neonate Screening
| Instrument | Type | Manufacturer | Order number |
|---|---|---|---|
| Micro Filter Plates | Multiscreen 96 well | Merck Millipore | MSHVN4550 |
| Microtiter Plates | 96 well Mikrotiter plates made of Polypropylen (F-Form) | Greiner | 655201 |
| Microtiter Plates Locks | Micromats | ||
| Aluminium Foil | |||
| Mass Spectrometer | Quattro Ultima (ESI-MS/MS) | Micromass (today: Waters) | |
| Methanol | (20837.320 METHANOL HIPERSOLV HPLC ISOCRATIC GRADE) |
Lab Materials
| Material | Name | Manufacturer |
|---|---|---|
| Bottle 50 ml, 100 ml, 200 ml, 500 ml, 1000 ml | ||
| Beaker 50 ml, 100 ml, 200 ml, 500 ml 1000 ml | ||
| Conical Flask 500 ml, 1000 ml, 100 ml | ||
| Conical Flask 300 ml | ||
| Single Channel Pipette | Pipetman® P2, P20, P200, P1000 | Gilson |
| Multichannel Pipette | ||
| Multistep Pipette | ||
| Pipette | ||
| 0.2 ml PCR Tube, Flat Cap, Natural | I1402-8200 | Starlab GmbH |
| Microcentrifuge Tubes | ||
| Centrifugetubes | ||
| PCR 8-Strips | ||
| Gloves Nitril Gr. L | Starlab powderfree | Starlab |
| Gloves Nitril Gr. M | Starlab powderfree | Starlab |
| Gloves Nitril Gr. S | Starlab powderfree | Starlab |
| Filtertips | ||
| Inoculating Loop | ||
| Disposal Bags | ||
| Flasks | ||
| Plate | ||
| Petri Dishes | P5731-500EA | Sigma-Aldrich Chemie GmbH |
| Petri Dishes | N221.2 | Carl Roth GmbH & Co.KG |
| TipOne® Pipette Tip | S1110-1700 | Starlab GmbH |
| TipOne® Pipette Tip, 1000μl, Graduated | S1111-2721 | Starlab GmbH |
| NeoBox-81 6er Set, je 1 x Transparent, g | 22916 | |
| NeoLab-Marker for Reaction-Flaks | 19079 | |
| Gene Pulser/MicroPulser Cuvettes, 0.1 cm | 165-2089 | |
| NeoLab-Paper Scissors, 23 cm Long, Even | 23272 | |
| TipOne® Pipette Tip, 200μl | S1110-1700 | Starlab GmbH |
| TipOne® Pipette Tip, 10μl, Graduated, Re | S1111-3700 | Starlab GmbH |
| Pipette, 5 ml | 606180 | Greiner bio-one GmbH |
| Ring out of Plumbum with Vinyl Coating, 57 mm In | 310161013 | NEOLAB GmbH |
| TipOne® Pipette Tip 10μl, Graduated, Rac | S1111-3800 | Starlab GmbH |
| Reaction Tube,S.L.1.5 ml,Colorless. | 12682 | Eppendorf, Fisher Scientific GmbH |
| Reaction Tube,S.L.,2 ml, Colorless | 12776 | Eppendorf |
| NeoTape-Writing Tape, 13 mm, Gray | 280126114 | NEOLAB GMBH |
| NeoTape-Writing Tape, 25 mm, Salmon-Colored | 280126229 | NEOLAB GMBH |
| 10 ml Serological Pipette, Filter, Sterile | E4860-1011 | Starlab GmbH |
| Gloves Latex + Alovera L | 14089 | |
| Gloves Latex + Alovera M | 14088 | |
| Gloves Latex + Alovera S | 14087 | |
| PH-Stripes | WHAT10362000 | PANPEHA PLUS |
| 100 Run24Barcode | 20110099-100 | GATC Biotech AG |
| Tube Conical, Polypropylen, 50 ml | 352070 | NEOLAB GMBH |
| Gene Pulser/MicroPulser Cuvettes, 0.1 cm | 165-2089 | Bio-Rad Laboratories GmbH |
| 0.2 ml 8-Tube Strips Without Caps, Nature | TBS-0201 | Bio-Rad Laboratories GmbH |
| Round-Bottom Tubule, Polypropylen, 14 ml | 352059 | NEOLAB GMBH |
| Optical Flat 8-Cap Strips | TCS-0803 | Bio-Rad Laboratories GmbH |
| Inoculation Loop 10 μl, Blue, Sterile | 2900254437 | |
| Corning Serological Pipette 50 ml | 14303 | |
| Weighing Dish 500 ST | 1884.1 | Carl Roth GmbH & Co.KG |
| Filter Paper | Z274836-1PAK | Sigma-Aldrich Chemie GmbH |
| Inoculation Loop 10 µl | 2900254437 | NEOLAB GMBH |
Further Recipies and Stocks
Acidovorans Complex Medium
for 1 L
Ampicillin Stock Solution
| Stock | Ampicillin 100 mg / ml |
| Amount | 50 ml |
| Storage | -24°C freezer |
| Notes | Use in 1:1000 dilution |
Efficiency Test

Figure 1B: Ampicilin efficiency test: Our ampicilin stock and a different ampicilin stock from the 3rd floor was diluted in 300 µl LB media, oculated with DH10beta + pSB1A6 (FannyTest plate, 2013-08-15) and grown at 37 °C for around 1.5 day. Additionally two LB media from the fridge already prepared with resistence were tested.
Bacitracin Stock Solution
| Stock | Bacitracin 5000 U/ml |
| Amount | 5 ml |
| Storage | -24°C freezer |
Brevibacillus Parabrevis Glycerol Stock
| Stock | Brevibacillus parabrevis ATCC8185 glycerol stock |
| Amount | 16 x 1.3 ml |
| Storage | -80°C freezer |
Chloramphenicol Stock Solution
| Stock | Chloramphenicol 30 mg / ml |
| Amount | 10 ml |
| Storage | -24°C freezer |
| Notes | Solved in 100% ethanol. Use in 1:3000 dilution |
D. Acidovorans Glycerol Stock
| Stock | D. acidovorans glycerol stock |
| Amount | 10 x 1 ml |
| Storage | -80°C freezern |
E.Coli BAP 1 Glycerol Stock
| Stock | E. coli BAP1 glycerol stock |
| Amount | 2 x 1.3 ml |
| Storage | -80°C freezer |
E.Coli BAP1, Competent
| Stock | E. coli BAP1 |
| Amount | 18 x 100 µl |
| Storage | -80°C freezer |
| Notes | Grows extremely fast. Be careful with miniPreps, at least in cultures with ampicillin it tends to degrade all available ampicillin and then lose the respective plasmid. |
E. coli BAP1-pKD46 Glycerol Stock
| What | E. coli BAP1-pKD46 glycerol stock (Ampr) |
| Amount | 2 x 1.3 ml |
| Storage | -80°C freezer |
| Notes | Grow at 30°C only! Growth at 37°C will lead to loss of pKD46 plasmid. |
E.Coli BAP1-pLF03 Glycerol Stock
| Stock | E. coli BAP1-pLF03 glycerol stock (Ampr) |
| Amount | 1 x 1.3 ml |
| Storage | -80°C freezer |
| Notes | Might have low amount of plasmid-carrying bacteria due to long culturing time (all Amp in medium cleaved) |
E. Coli DH5α-pCP20 Glycerol Stock
| Stock | E. coli DH5α-pCP20 glycerol stock (Ampr, Cmr) |
| Amount | 2 x 1.3 ml |
| Storage | -80°C freezer |
| Notes | Grow at 30°C only! Growth at 37°C will lead to loss of pCP20 plasmid. |
Heat-Shock Competent E. Coli TOP10
| Stock | E. coli TOP10, Heat-shock competent |
| Amount | 200 x 100 µl |
| Storage | -80°C freezer |
| Notes | Verified on 2013-06-07. |

E. Coli TOP10-BBa I746200/pSB1AK3 Glycerol Stock
| Stock | E. coli TOP10-(BBa I746200 in pSB1AK3) (Ampr, Kanr) |
| Amount | 1 x 1.3 ml |
| Storage | -80°C freezer |
E. Coli TOP10-BBa J04450/pSB3C5 Glycerol Stock
| Stock | E. coli TOP10-(BBa_J04450 in pSB3C5) glycerol stock (Cmr) |
| Amount | 1 x 1.3 ml |
| Storage | -80°C freezer |
E. Coli TOP10-pIK1 Glycerol Stock
| Stock | E. coli TOP10-pIK1 glycerol stock (Cmr) |
| Amount | 1 x 1.3 ml |
| Storage | -80°C freezer |
E. Coli TOP10-pKD46 Glycerol Stock
| Stock | E. coli TOP10-pKD46 glycerol stock (Ampr) |
| Amount | 2 x 1.3 ml |
| Storage | -80°C freezer |
| Notes | Grow at 30°C only! Growth at 37°C will lead to loss of pKD46 plasmid. |
E. Coli TOP10-pMM65 Glycerol Stock
| Stock | E. coli TOP10-pMM65 glycerol stock (Kanr) |
| Amount | 1 x 1.3 ml |
| Storage | -80°C freezer |
IPTG Stock Solution
| Stock | IPTG 23.8 mg / ml |
| Amount | 10 ml |
| Storage | -20°C freezer |
| Notes | Use 0.1 to 1 mM |
Kanamycin Stock Solution
| Stock | Kanamycin 50 mg / ml |
| Amount | 10 ml |
| Storage | -24°C freezer |
| Notes | Use in 1:1000 dilution |
M9 Medium
| Name | M9 Minimal Salts 5x, Powder |
| Amount | 1 l |
| Storage | room temperature |
| Notes | close tightly, hygroscopic |
Reactivation Medium
for 1L
Solution for MS Preparation
Solution 1 (for MS Sample Preparation Neo Gen Stable Isotope Standard Kit A and B (NSK-AB, EurIsotop)
1. One vial of amino acid standard (NSK-A) is taken up in 1 ml methanol/water (1:1 v/v) and solubilized for ca. 15 minutes in an ultra-sonification bath. Caution: make sure that the lid is closed well and not wetted by water of the ultra-sonification bath.
2. One vial of acylcarnitine standard (NSK-B) is taken up in 1 ml of methanol and solubilized for ca. 15 minutes in the ultra-sonification bath.Caution: make sure that the lid is closed well and not wetted by water of the ultra-sonification bath.
Contents of both vials are transfered quantitatively with methanol in a 200 ml graduate flask.The graduate flask is filled up to the calibration mark with methanol.
Standard solutions get a consecutive number. When two 200 ml graduate flasks are prepared simultaneously, contents of both flasks can be combined, if levels of both standards (old and new) coincide. Afterwards the whole solution will be labeled with one common number.
Solution 2 (for MS sample preparation): butanolic hydrochloric acid (3 N), water-free
1. 900 ml butanol (p.A., Merck 1.01990.1000) are cooled in an ice bath at 0°C (magnetic stirrer).
The vessel has to be closed to avoid ice condensation of water.
2. 100 ml acetylchloride (p.A., Merck 1.00031.0250) are added dropwiese from a dropping funnel over a a period of 5 minutes while stirring constantly (protection goggles and gloves!).
Caution: make sure that no water or ice gets into the solution.
3. The solution is stirred for additional 10 minutes with closed lid on ice.
Solution 3 (for MS sample preparation)
Acetonitrile p.A./water (1:1 v/v) (83639.320DE ACETONITRIL HIPERSOLV SUPER GR REAG.PE)
 "
"









