Team:Macquarie Australia/Project
From 2013.igem.org




General
A seperate
Overall project
The iGEM team at Macquarie University are aiming to introduce the genes necessary for chlorophyll production into E.coli. We at Macquarie are confident that we can make scientific strides in the understanding and construction of a photosynthetic bacterium.
Currently we have 2 out of 12 biobricks assembled and sequenced in our system, with work continuing on the remainder.
Production of chlorophyll in E.coli would be the first steps towards the construction of photosystem II, a fundamental aspect of organic energy production. A better understanding of photosystem II opens the door to the production of harnessing green energy.
If successful this would be the first successful production of chlorophyll within non-photosynthetic bacteria.
Aims
Chlorophyll Biosynthesis Gene Pathway

Chll1 - Magnesium chelatase subunit I
Forms an ATP dependent hexameric ring complex and a complex with the ChlD subunit (probably a double hexameric ring complex) before acting on the protoporphyrin which is bound to the ChlH protein to insert magnesium [PMID: 11469861]. Transcript is light regulated and may be diurnal and/or circadian [PMID: 16228385]; predicted chloroplast targeting sequence amino acids 1-54 by ChloroP.
Chll2 - Magnesium chelatase subunit I
forms an ATP dependent hexameric ring complex and a complex with the ChlD subunit (probably a double hexameric ring complex) before acting on the protoporphyrin which is bound to the ChlH protein to insert magnesium [PMID: 11469861]; may have similar function to Arabidopsis CHLI2 gene [PMID: 11842180]; chloroplast targeting signal peptide predicted 1-37 by ChloroP.
ChlD - Magnesium chelatase subunit D
Forms an ATP dependent complex with the ChlI subunit (probably a double hexameric ring complex) before acting on the protoporphyrin which is bound to the ChlH protein to insert magnesium [PMID: 11469861]. Predicted chloroplast targeting sequence amino acids 1-62 by ChloroP.
ChlH - Magnesium chelatase subunit H
chloroplast precursor; Chlamydomonas mutants with defects in this protein are chl1 and brs-1 and result in a brown phenotype [PMID: 11713666; PMID: 4436384]. Orthologous to the bacterial protein BchH [PMID: 9359397]; binds protoporphyrin and is acted upon by the ChlI:ChlD complex for magnesium insertion [PMID: 11469861]; interacts with GUN4 and may be involved in chloroplast signalling: Gene is also known as GUN5 in Arabidopsis thaliana [PMID: 11172074; 12574634]; transcript is light regulated and may be diurnal and/or circadian regulated [PMID: 16228385].
Gun4 - Tetrapyrrole-binding protein
In Arabidopsis, GUN4 (Genomes uncoupled 4) is required for the functioning of the plastid mediated repression of nuclear transcription that is involved in controlling the levels of magnesium- protoporphyrin IX. GUN4 binds the product and substrate of Mg-chelatase, an enzyme that produces Mg-Proto, and activates Mg-chelatase. GUN4 is thought to participates in plastid-to-nucleus signaling by regulating magnesium-protoporphyrin IX synthesis or trafficking.
ChlM - Mg protoporphyrin IX S-adenosyl methionine O-methyl transferase
Magnesium-protoporphyrin O-methyltransferase (chlM) [PMID: 12828371; PMID: 12489983; PMID: 4436384]; ChloroP 1.1 predicts cp location
CTH1 - Copper target 1 protein
functional variant produced under copper and/or oxygen sufficient conditions [GI:15650866; PMID: 11910013; PMID: 14673103]; CTH1; Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase, aerobic oxidative cyclase; orthologous to Rubrivaxgelatinosus aerobic oxidative cyclase [PMID: 11790744; PMID: 14617630]; predicted chloroplast transit peptide 1-35; Orthologous to CRD1; CHL27B [PMID: 15849308].
Plastocyanin - Chloroplast precursor
pre-apoplastocyanin, PETE [PMID: 2165059; PMID: 8940133]; structure of plastocyanin PDB: 2PLT; mutant = ac208 [PMID: 8463310]
POR - Light-dependent protochlorophyllidereductase
Light-dependent protochlorophyllidereductase, chloroplast precursor; Converts protochlorophyllide to chlorophyllide using NADPH and light as the reductant; Chlamydomonas mutant known as pc-1 has a two-nucleotide deletion within the fourth and fifth codons of this gene giving rise to a premature termination [PMID: 8616232; identical to U36752]
DVR1 - 3,8-divinyl protochlorophyllidea 8-vinyl reductase
Predicted chloroplast transit peptide 1-58; [PMID: 15695432; PMID: 15849308]
ChlG - Chlorophyllsynthetase
Catalyses the esterification of chlorophyllide with phytyl-pyrophosphate to make chlorophyll
ChlP - Geranylgeranyl reductase
Reduces the geranylgeranyl group to the phytyl group in the side chain of chlorophyll. Plant geranylgeranylhydrogenase (CHL P) reduces free geranylgeranyldiphosphate to phytildiphosphate, which provides the side chain to chlorophylls, tocopherols, and plastoquinones.
Methods and workflow
Design
We designed 10 genes necessary for chlorophyllide biosynthesis and another 2 genes for chlorophyll biosynthesis, totaling 12 genes. These genes were also codon optimised for expression within E. coli.
Assembly
Using Gibson Assembly we can reassemble our genes insert them into the plasmid backbone. This removes the need for ligations and restriction digests. Allowing the production of complete BioBricks without the need for extra steps to get the gene into the destination plasmid.
Transformation
By transforming in E. coli we can determine if the gene is functional as well as purify the plasmid. By transforming in top10 strain E. coli we can overproduce the proteins and then characterise the BioBricks produced.
Sequence
It is imperative that the plasmids produced from the Gibson Assembly be sequenced to determine if there have been any nucleotide changes between the planned sequences and those synthesised. Therefore sequencing data needs to be gathered before any ligations are performed to ensure the correct construction of our gene pathway. This will also demonstrate that the protein sequence has not changed and the protein should therefore be functional.
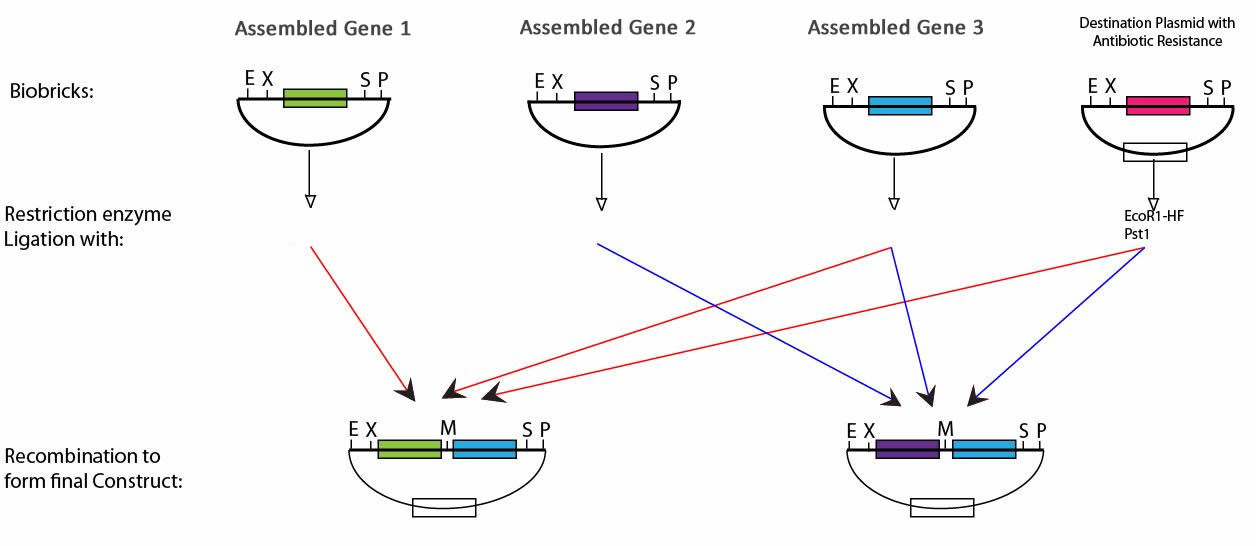
BioBrick Assembly
Following digestion of the BioBricks produced with the appropriate enzyme and ligation it is possible to produce the plasmids required for chlorophyll biosynthesis. This protocol can be seen below,
Transformation & Characterizations
After ligating BioBricks to assemble our gene pathway we will be able to show the usefulness of Gibson Assembly in synthetic biology. This will provide a means to characterise the two biobricks simultaneously.
Highlighted results
[Part 2-The Experiments-Part 3-Results]
 "
"