Team:Edinburgh/Introduction/Metal promoters
From 2013.igem.org
Hristianita (Talk | contribs) |
Krothschild (Talk | contribs) |
||
| (14 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div class='content'> | <div class='content'> | ||
| + | <h3>Fur transcription factor and the fur box </h3> | ||
| + | |||
| + | |||
| + | In <i>Bacillus subtilis</i>, iron recognition is achieved via the Ferric Uptake Regulator (Fur). Fur normally forms a dimer and interacts with a specific operator called the fur box to repress genes further downstream. At high intra-cellular iron concentrations, Fur binds to iron to form a holo-repressor and then binds to the fur box and represses gene transcription. Genes controlled by the fur box usually code for iron binding proteins and gate mechanisms within the cell (about 40 genes are controlled by the fur box). In the absence of significant iron levels, the apo-repressor Fur does not bind to the fur box and the relevant genes can be coded for. | ||
| + | |||
| + | |||
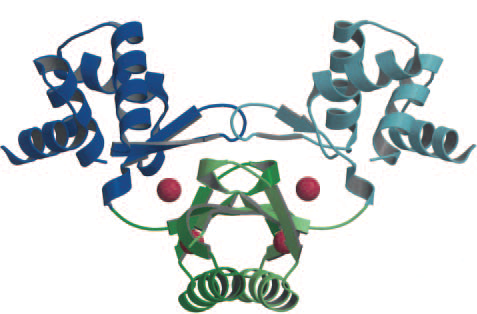
| + | There are two iron binding sites per Fur monomer, so 4 overall sites overall in the dimer. (Pohl et al., 2003) The upper binding site (S2) regularly has a zinc ion for structural integrity. The lower binding site (S1) typically has a zinc ion, which is replaced by iron when the intracellular iron concentration rises (Figure 1). | ||
| + | |||
| + | |||
| + | |||
| + | [[File:fur.jpg.png]] | ||
| + | |||
| + | '' '''Figure 1.''' ''(Pohl et al., 2003). The Fur dimer crystal structure bound to 4 zinc ions (red spheres). The two upper zinc ion are for structural integrity, the two lower zinc ions are for iron capture (replaced by iron when concentrations are high enough). | ||
| + | |||
| + | |||
| + | The fur box pattern that is conserved in all different fur boxes is the palindromic sequence: TGATAAT-N-ATTATCA (N being any base). Most fur boxes are 19 to 21 base pairs and always contain this 7-1-7 palyndromic sequence. Different theories exist as to how the Fur dimer binds to the fur box. The classical model states that a Fur dimer binds to one fur box and represses the downstream gene. Another states that the hexameric GATAAT pattern is recognized by three different fur dimers (Fuangthong and Helmann, 2003). The most recent one posits that the minimal binding site for the Fur dimer is TGATAAT-N-ATTATCA and that most fur boxes contain two repeats of this pattern, such as TGATAATGATAATCATTATCA (Figure 2). | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Boii.png]] | ||
| + | |||
| + | '' '''Figure 2.''' '' The most recent study (Baichoo and Helmann, 2002) suggest most fur boxes have two overlapping palindromic sequence and two fur dimers bind. | ||
| + | |||
| + | |||
| + | |||
| + | We decided to assess the minimal binding site for the fur box to work (TGATAAT-N-ATTATCA) in a synthetic construct and assay the effect of iron on a fluorescent protein located downstream. | ||
| + | |||
| + | |||
| + | |||
| + | <h2>References </h2> | ||
| + | |||
| + | Baichoo, N. and Helmann, J. (2002) Recognition of DNA by Fur: a Reinterpretation of the Fur Box Consensus Sequence. Jour. of Bacter. 184, 5826-5832. | ||
| + | |||
| + | Fuangthong, M. and Helmann J. (2003) Recognition of DNA by Three Ferric Uptake Regulator Homologs in Bacillus Subtilis. Jour. of Bacter. 185, 6348-6357. | ||
| + | |||
| + | Pohl, E., Haller, J., Mijovilovich, A., Meyer-Klaucke, W., Garman, E. and Vasil, M. (2003) Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Micr. 47 (4), 903-915. | ||
</div> | </div> | ||
{{Team:Edinburgh/Footer}} | {{Team:Edinburgh/Footer}} | ||
Latest revision as of 01:10, 5 October 2013
Fur transcription factor and the fur box
In Bacillus subtilis, iron recognition is achieved via the Ferric Uptake Regulator (Fur). Fur normally forms a dimer and interacts with a specific operator called the fur box to repress genes further downstream. At high intra-cellular iron concentrations, Fur binds to iron to form a holo-repressor and then binds to the fur box and represses gene transcription. Genes controlled by the fur box usually code for iron binding proteins and gate mechanisms within the cell (about 40 genes are controlled by the fur box). In the absence of significant iron levels, the apo-repressor Fur does not bind to the fur box and the relevant genes can be coded for.
There are two iron binding sites per Fur monomer, so 4 overall sites overall in the dimer. (Pohl et al., 2003) The upper binding site (S2) regularly has a zinc ion for structural integrity. The lower binding site (S1) typically has a zinc ion, which is replaced by iron when the intracellular iron concentration rises (Figure 1).
Figure 1. (Pohl et al., 2003). The Fur dimer crystal structure bound to 4 zinc ions (red spheres). The two upper zinc ion are for structural integrity, the two lower zinc ions are for iron capture (replaced by iron when concentrations are high enough).
The fur box pattern that is conserved in all different fur boxes is the palindromic sequence: TGATAAT-N-ATTATCA (N being any base). Most fur boxes are 19 to 21 base pairs and always contain this 7-1-7 palyndromic sequence. Different theories exist as to how the Fur dimer binds to the fur box. The classical model states that a Fur dimer binds to one fur box and represses the downstream gene. Another states that the hexameric GATAAT pattern is recognized by three different fur dimers (Fuangthong and Helmann, 2003). The most recent one posits that the minimal binding site for the Fur dimer is TGATAAT-N-ATTATCA and that most fur boxes contain two repeats of this pattern, such as TGATAATGATAATCATTATCA (Figure 2).
Figure 2. The most recent study (Baichoo and Helmann, 2002) suggest most fur boxes have two overlapping palindromic sequence and two fur dimers bind.
We decided to assess the minimal binding site for the fur box to work (TGATAAT-N-ATTATCA) in a synthetic construct and assay the effect of iron on a fluorescent protein located downstream.
References
Baichoo, N. and Helmann, J. (2002) Recognition of DNA by Fur: a Reinterpretation of the Fur Box Consensus Sequence. Jour. of Bacter. 184, 5826-5832.
Fuangthong, M. and Helmann J. (2003) Recognition of DNA by Three Ferric Uptake Regulator Homologs in Bacillus Subtilis. Jour. of Bacter. 185, 6348-6357.
Pohl, E., Haller, J., Mijovilovich, A., Meyer-Klaucke, W., Garman, E. and Vasil, M. (2003) Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Micr. 47 (4), 903-915.

| 
| | | | 
|
| This iGEM team has been funded by the MSD Scottish Life Sciences Fund. The opinions expressed by this iGEM team are those of the team members and do not necessarily represent those of MSD | |||||
 "
"