Team:Paris Saclay/Notebook/August/29
From 2013.igem.org
(→2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI) |
(→2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI) |
||
| (30 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
===='''Objective : characterize BBa_K1155000 and BBa_K1155004, BBa_K1155005, BBa_K1155006'''==== | ===='''Objective : characterize BBa_K1155000 and BBa_K1155004, BBa_K1155005, BBa_K1155006'''==== | ||
| - | ===='''1 - Purification of | + | ===='''1 - Purification of colonies: NirB with RBS-LacZ-Term in pSB1C3, Pfnr with RBS-AmilCP-Term in pSB1C3 by streaking colonies in aerobic or anaerobic conditions'''==== |
XiaoJing | XiaoJing | ||
| Line 15: | Line 15: | ||
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DE;" | | | style="border:1px solid black;padding:5px;background-color:#DE;" | | ||
| - | Purification of 08/28/13 didn't work. We have blue colonies for | + | Purification of 08/28/13 didn't work. We have blue colonies for Pndh* with RBS-AmilCP-Term in pSB1C3 in aerobic and anaerobic conditions. We also have blue colonies for NirB with RBS-LacZ-Term in pSB1C3 in anaerobic conditions. We will streak these colonies again. |
|} | |} | ||
| Line 21: | Line 21: | ||
[[File:PsNirB2908.jpg|500px]] | [[File:PsNirB2908.jpg|500px]] | ||
| - | We streak : | + | We streak colonies from construction : |
| - | * | + | * Pndh* with RBS-Amil CP-Term in pSB1C3 with O2 at 37°C |
| - | * | + | * Pndh* with RBS-Amil CP-Term in pSB1C3 without O2 at 37°C |
| - | * | + | * Pndh* with RBS-Amil CP-Term in pSB1C3 with O2 at 30°C |
| - | * | + | * Pndh* with RBS-Amil CP-Term in pSB1C3 without O2 at 30°C |
* NirB with RBS-LacZ-Term in pSB1C3 with O2 with Xgal at 37°C | * NirB with RBS-LacZ-Term in pSB1C3 with O2 with Xgal at 37°C | ||
* NirB with RBS-LacZ-Term in pSB1C3 without O2 with Xgal at 37°C | * NirB with RBS-LacZ-Term in pSB1C3 without O2 with Xgal at 37°C | ||
| - | We also purify | + | We also purify Pndh* with RBS-AmilCP-Term in pSB1C3 in liquid culture at 37°C using : |
| - | * | + | * Pndh* with RBS-AmilCP-Term in pSB1C3 in aerobic conditions : |
** LB : 10 mL | ** LB : 10 mL | ||
** Clone : 1 and 2 | ** Clone : 1 and 2 | ||
| - | * | + | * Pndh* with RBS-AmilCP-Term in pSB1C3 in anaerobic conditions : |
** LB : 50mL | ** LB : 50mL | ||
** Clone : 1 and 2 | ** Clone : 1 and 2 | ||
| - | ===='''2 - | + | ===='''2 - PCR Colony of strain DH5α containing plasmid pSB1C3 with Pndh* and RBS_AmilCP-Term'''==== |
XiaoJing | XiaoJing | ||
| - | We | + | We resuspend our colonies in 20µL of H2O. |
Used quantities : | Used quantities : | ||
| - | * DNA : 2µL | + | * DNA : 2µL of resuspend colony |
| - | * | + | * PCR mix: 23µL |
| + | |||
| + | PCR mix: | ||
** Oligo 43 : 14µL | ** Oligo 43 : 14µL | ||
** Oligo 44 : 14µL | ** Oligo 44 : 14µL | ||
| Line 78: | Line 80: | ||
** H2O : 9µL | ** H2O : 9µL | ||
| - | We | + | We incubatethe digestion for 30 minutes at 37°C. |
===='''4 - Electrophoresis to check the digestion of BBa_K1155000 by PstI/SpeI, BBa_K1155003, BBa_K1155007 by Xbal/PstI'''==== | ===='''4 - Electrophoresis to check the digestion of BBa_K1155000 by PstI/SpeI, BBa_K1155003, BBa_K1155007 by Xbal/PstI'''==== | ||
| Line 85: | Line 87: | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" | [[]] | + | | style="width:350px;border:1px solid black;" | [[File:Psgel12908.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL DNA Ladder | * Well 1 : 6µL DNA Ladder | ||
| - | * Well 2 : 5µL of RBS- | + | * Well 2 : 5µL of RBS-AmilCP-Term clone 9 + 1µL of 6X loading dye |
| - | * Well 3 : 5µL of RBS- | + | * Well 3 : 5µL of RBS-AmilCP-Term clone 12 + 1µL of 6X loading dye |
| - | * Well 4 : 5µL of RBS-LacZ-Term clone 10+1µL of 6X loading dye | + | * Well 4 : 5µL of RBS-LacZ-Term clone 10 + 1µL of 6X loading dye |
| - | * Well 5 : 5µL of RBS-LacZ-Term clone 11+1µL of 6X loading dye | + | * Well 5 : 5µL of RBS-LacZ-Term clone 11 + 1µL of 6X loading dye |
| - | * Well 6 : 5µL of RBS-LacZ-Term clone 15+1µL of 6X loading dye | + | * Well 6 : 5µL of RBS-LacZ-Term clone 15 + 1µL of 6X loading dye |
* Gel : 1.0% | * Gel : 1.0% | ||
|} | |} | ||
| Line 98: | Line 100: | ||
Expected sizes : | Expected sizes : | ||
* RBS-LacZ-Term : 3500bp | * RBS-LacZ-Term : 3500bp | ||
| - | * RBS- | + | * RBS-AmilCP-Term : 824bp |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Expected sizes : | Expected sizes : | ||
| - | * | + | * Pndh* : 111bp |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We | + | We obtained fragments at the right size. We will purify them. |
|} | |} | ||
| Line 123: | Line 117: | ||
Nanodrop : | Nanodrop : | ||
| - | * | + | * Pndh* : 26.2ng/µL |
===='''6 - Electrophoresis to check the gel purification of the digestion of BBa_K1155000 by PstI/SpeI, BBa_K1155003, BBa_K1155007 by Xbal/PstI'''==== | ===='''6 - Electrophoresis to check the gel purification of the digestion of BBa_K1155000 by PstI/SpeI, BBa_K1155003, BBa_K1155007 by Xbal/PstI'''==== | ||
| Line 130: | Line 124: | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" | [[]] | + | | style="width:350px;border:1px solid black;" | [[File:Psgel32908.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL DNA Ladder | * Well 1 : 6µL DNA Ladder | ||
| - | * Well 2 : 2µL of RBS-LacZ-Term clone 10+1µL of 6X loading dye | + | * Well 2 : 2µL of RBS-LacZ-Term clone 10 + 1µL of 6X loading dye |
| - | * Well 3 : 2µL of RBS- | + | * Well 3 : 2µL of RBS-AmilCP-Term clone 9 + 1µL of 6X loading dye |
* Gel : 1.0% | * Gel : 1.0% | ||
|} | |} | ||
| Line 140: | Line 134: | ||
Expected sizes : | Expected sizes : | ||
* RBS-LacZ-Term : 3500bp | * RBS-LacZ-Term : 3500bp | ||
| - | * RBS-Amil CP-Term : | + | * RBS-Amil CP-Term : 824bp |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We | + | We obtained fragments at the right size for RBS-LacZ-Term. We will ligate it. |
|} | |} | ||
| Line 152: | Line 146: | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" | [[]] | + | | style="width:350px;border:1px solid black;" | [[File:Psgel42908.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL DNA Ladder | * Well 1 : 6µL DNA Ladder | ||
| - | * Well 2 : 5µL of | + | * Well 2 : 5µL of PNirB clone 7 + 1µL of 6X loading dye |
| - | * Well 3 : 5µL of NarG clone 6+1µL of 6X loading dye | + | * Well 3 : 5µL of NarG clone 6 + 1µL of 6X loading dye |
| - | * Well 4 : 5µL of BBa_K1155006 already digested by SpeI+1µL of 6X loading dye | + | * Well 4 : 5µL of BBa_K1155006 already digested by SpeI + 1µL of 6X loading dye |
| - | * Well 5 : 5µL of BBa_K1155004 already digested by SpeI+1µL of 6X loading dye | + | * Well 5 : 5µL of BBa_K1155004 already digested by SpeI + 1µL of 6X loading dye |
| - | * Well | + | * Well 6 : 5µL of BBa_K1155005 already digested by SpeI + 1µL of 6X loading dye |
* Gel : 1.0% | * Gel : 1.0% | ||
|} | |} | ||
Expected sizes : | Expected sizes : | ||
| - | * NarK, NarG, NirB : | + | * NarK, NarG, NirB : 2000bp |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We | + | We obtained fragments at the right size for NarK. We will ligate it |
| - | |} | + | |} |
| + | |||
| + | ===='''8 - Purification colony of strain MG1655Z1 Δfnr '''==== | ||
| + | |||
| + | XiaoJing | ||
| + | |||
| + | {| | ||
| + | | style="border:1px solid black;padding:5px;background-color:#DE;" | | ||
| + | Culture from 08/28/13 works. We will do a purification of one colonies on plates with kanamycin, ampicilin and chloramphenicol antibiotics. | ||
| + | |} | ||
| + | |||
| + | We streaked 4 colonies in each plate but we used the same colony to streak on plates with LB or kanamycin or ampicilin or chloramphenicol antibiotics. We incubate our colonies at 42°C. | ||
===='''Objective : obtaining Pfnr, NarK, NarG or NirB and RBS-LacZ-Term or RBS-AmilCP-Term in pSB3K3'''==== | ===='''Objective : obtaining Pfnr, NarK, NarG or NirB and RBS-LacZ-Term or RBS-AmilCP-Term in pSB3K3'''==== | ||
| Line 185: | Line 190: | ||
* PstI FD : 1µL | * PstI FD : 1µL | ||
| - | We | + | We incubate the digestion at 37°C for 10 minutes. |
===='''2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI'''==== | ===='''2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI'''==== | ||
| Line 192: | Line 197: | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" | [[]] | + | | style="width:350px;border:1px solid black;" | [[File:Psgel52908.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL DNA Ladder | * Well 1 : 6µL DNA Ladder | ||
| - | * Well 3 : 5µL of PSB3K3+1µL of 6X loading dye | + | * Well 3 : 5µL of PSB3K3 + 1µL of 6X loading dye |
* Gel : 1.0% | * Gel : 1.0% | ||
|} | |} | ||
| Line 203: | Line 208: | ||
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We | + | We obtained fragments at the right size. We will ligate it. |
|} | |} | ||
| Line 213: | Line 218: | ||
Nanodrop : | Nanodrop : | ||
| - | * | + | * Pndh* : 7.2ng/µL |
| - | + | ||
==='''A - Aerobic/Anaerobic regulation system / B - PCB sensor system'''=== | ==='''A - Aerobic/Anaerobic regulation system / B - PCB sensor system'''=== | ||
| Line 220: | Line 224: | ||
===='''Objective : obtaining FRN and BphR2 proteins'''==== | ===='''Objective : obtaining FRN and BphR2 proteins'''==== | ||
| - | ===='''1 - | + | ===='''1 - PCR Colony of FNR, RBS-FNR and RBS-BphR2 in DH5α strain'''==== |
XiaoJing | XiaoJing | ||
Latest revision as of 01:22, 5 October 2013
Notebook : August 29
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000 and BBa_K1155004, BBa_K1155005, BBa_K1155006
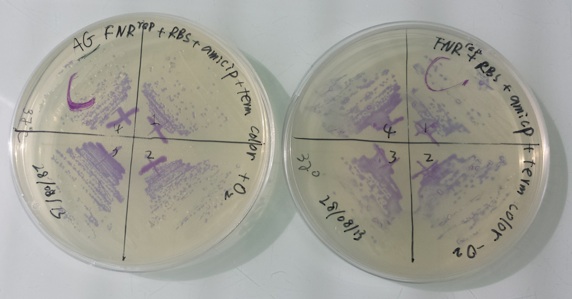
1 - Purification of colonies: NirB with RBS-LacZ-Term in pSB1C3, Pfnr with RBS-AmilCP-Term in pSB1C3 by streaking colonies in aerobic or anaerobic conditions
XiaoJing
|
Purification of 08/28/13 didn't work. We have blue colonies for Pndh* with RBS-AmilCP-Term in pSB1C3 in aerobic and anaerobic conditions. We also have blue colonies for NirB with RBS-LacZ-Term in pSB1C3 in anaerobic conditions. We will streak these colonies again. |
We streak colonies from construction :
- Pndh* with RBS-Amil CP-Term in pSB1C3 with O2 at 37°C
- Pndh* with RBS-Amil CP-Term in pSB1C3 without O2 at 37°C
- Pndh* with RBS-Amil CP-Term in pSB1C3 with O2 at 30°C
- Pndh* with RBS-Amil CP-Term in pSB1C3 without O2 at 30°C
- NirB with RBS-LacZ-Term in pSB1C3 with O2 with Xgal at 37°C
- NirB with RBS-LacZ-Term in pSB1C3 without O2 with Xgal at 37°C
We also purify Pndh* with RBS-AmilCP-Term in pSB1C3 in liquid culture at 37°C using :
- Pndh* with RBS-AmilCP-Term in pSB1C3 in aerobic conditions :
- LB : 10 mL
- Clone : 1 and 2
- Pndh* with RBS-AmilCP-Term in pSB1C3 in anaerobic conditions :
- LB : 50mL
- Clone : 1 and 2
2 - PCR Colony of strain DH5α containing plasmid pSB1C3 with Pndh* and RBS_AmilCP-Term
XiaoJing
We resuspend our colonies in 20µL of H2O.
Used quantities :
- DNA : 2µL of resuspend colony
- PCR mix: 23µL
PCR mix:
- Oligo 43 : 14µL
- Oligo 44 : 14µL
- dNTP : 14µL
- Buffer Dream Taq : 69µL
- Dream Taq : 5.5µL
- H2O : 577µL
3 - Digestion of BBa_K1155000, BBa_K1155003, BBa_K1155007 by Xbal/PstI
XiaoJing
We used clone 9 and 12 for BBa_K1155003, clone 10, 11 and 15 for BBa_K1155007
Used quantities :
- BBa_K1155003, BBa_K1155007
- DNA : 14µL
- Buffer FD : 2µL
- XbaI FD : 2µL
- PstI FD : 2µL
- BBa_K1155000 :
- DNA : 5µL
- Buffer FD : 2µL
- SpeI FD : 2µL
- PstI FD : 2µL
- H2O : 9µL
We incubatethe digestion for 30 minutes at 37°C.
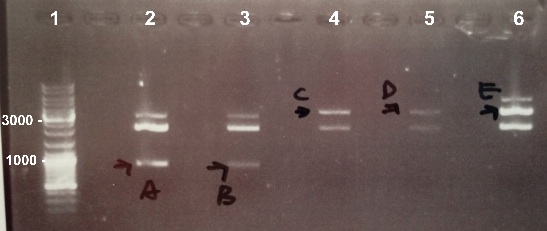
4 - Electrophoresis to check the digestion of BBa_K1155000 by PstI/SpeI, BBa_K1155003, BBa_K1155007 by Xbal/PstI
XiaoJing
Expected sizes :
- RBS-LacZ-Term : 3500bp
- RBS-AmilCP-Term : 824bp
Expected sizes :
- Pndh* : 111bp
|
We obtained fragments at the right size. We will purify them. |
5 - Gel purification of the digestion of BBa_K1155000 by PstI/SpeI, BBa_K1155003, BBa_K1155007 by Xbal/PstI
XiaoJing
Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ]
Nanodrop :
- Pndh* : 26.2ng/µL
6 - Electrophoresis to check the gel purification of the digestion of BBa_K1155000 by PstI/SpeI, BBa_K1155003, BBa_K1155007 by Xbal/PstI
Damir

|
|
Expected sizes :
- RBS-LacZ-Term : 3500bp
- RBS-Amil CP-Term : 824bp
|
We obtained fragments at the right size for RBS-LacZ-Term. We will ligate it. |
7 - Electrophoresis to check the digestion of BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI/PstI and plasmids already digested by SpeI and after digested by PstI
XiaoJing
Expected sizes :
- NarK, NarG, NirB : 2000bp
|
We obtained fragments at the right size for NarK. We will ligate it |
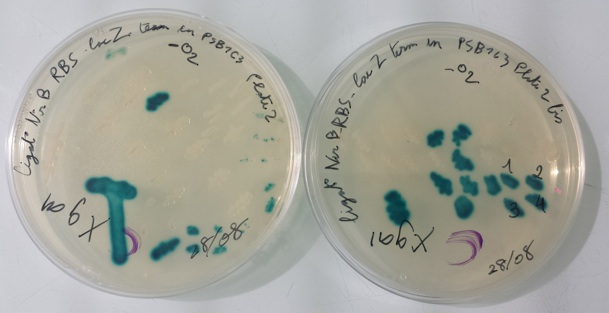
8 - Purification colony of strain MG1655Z1 Δfnr
XiaoJing
|
Culture from 08/28/13 works. We will do a purification of one colonies on plates with kanamycin, ampicilin and chloramphenicol antibiotics. |
We streaked 4 colonies in each plate but we used the same colony to streak on plates with LB or kanamycin or ampicilin or chloramphenicol antibiotics. We incubate our colonies at 42°C.
Objective : obtaining Pfnr, NarK, NarG or NirB and RBS-LacZ-Term or RBS-AmilCP-Term in pSB3K3
1 - Digestion of BBa_J04450 by EcoRI/PstI
Anaïs
Used quantities :
- Buffer FD: 2µL
- H2O : 5µL
- DNA : 9µL
- EcoRI FD : 1µL
- PstI FD : 1µL
We incubate the digestion at 37°C for 10 minutes.
2 - Electrophoresis to check the digestion of BBa_J04450 by EcoRI/PstI
XiaoJing
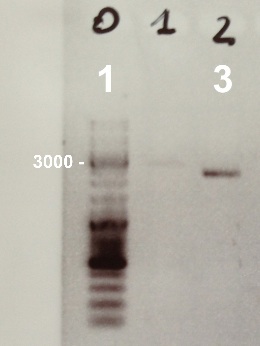
|
|
Expected sizes :
- pSB3K3 : 2750bp
|
We obtained fragments at the right size. We will ligate it. |
3 - Gel purification of the digestion of BBa_J04450 by PstI/SpeI
XiaoJing
Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ]
Nanodrop :
- Pndh* : 7.2ng/µL
A - Aerobic/Anaerobic regulation system / B - PCB sensor system
Objective : obtaining FRN and BphR2 proteins
1 - PCR Colony of FNR, RBS-FNR and RBS-BphR2 in DH5α strain
XiaoJing
|
Transformation of 08/28/13 works. We will do a Colony PCR. |
We mix our colonies in 20µL of H2O.
Used quantities :
- DNA : 2µL
- Mix : (it was divided in 8 tubes for 8 different colonies for each assembly with 23µL of mix in each tube. We do it twice.)
- Oligo 43 : 27.5µL
- Oligo 44 : 27.5µL
- dNTP : 27.5µL
- Buffer Dream Taq : 137.5µL
- Dream Taq : 11µL
- H2O : 1144µL
| Previous day | Back to calendar | Next day |
 "
"