|
|
| (68 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{Team:Uppsala/standard-style}} | | {{Team:Uppsala/standard-style}} |
| - | | + | {{Team:Uppsala/standard-style-no-frame}} |
| | | | |
| | <html> | | <html> |
| Line 10: |
Line 10: |
| | <link rel="stylesheet" type="text/css" href="css/style.css"> | | <link rel="stylesheet" type="text/css" href="css/style.css"> |
| | <style type="text/css"> | | <style type="text/css"> |
| | + | .hej{ |
| | + | width: 900px; |
| | + | } |
| | + | p *{text-align:justify;} |
| | | | |
| | </style> | | </style> |
| | + | |
| | + | <script type="text/javascript"> |
| | + | /*Acts once the document is loaded*/ $(document).ready(function() { /*Move the tn-main-wrap as child of <body>*/ $("#tn-main-wrap-wrap").prependTo($("body")); /*Move the menubars in the tn-main-wrap*/ $("#menubar.left-menu").appendTo($("#tn-main-wrap")); $("#menubar.right-menu").appendTo($("#tn-main-wrap")); |
| | + | /*Spoiler JS*/ $(".tn-spoiler div").slideUp(); $(".tn-spoiler a").click(function(e) { e.preventDefault(); $(".tn-spoiler-active").removeClass("tn-spoiler-active"); $(this).parent().addClass("tn-spoiler-active"); $(".tn-spoiler").not(".tn-spoiler-active").children("div").slideUp(); $(".tn-spoiler-active").children("div").slideToggle(); }); |
| | + | /*Add favicon*/ $("link[rel='shortcut icon']").remove(); $("head").append("<link rel='shortcut icon' type='image/png' href='https://static.igem.org/mediawiki/2013/6/6d/Uppsalas-cow-con.png'>"); }); |
| | + | </script> |
| | + | |
| | + | <link href="https://2013.igem.org/Team:Uppsala/lightbox-css-code.css?action=raw&ctype=text/css" type="text/css" rel="stylesheet"> |
| | + | |
| | + | <script src="https://2013.igem.org/Team:Uppsala/jquery-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> |
| | + | |
| | + | <script src="https://2013.igem.org/Team:Uppsala/lightbox-code.js?action=raw&ctype=text/javascript" type="text/javascript"></script> |
| | | | |
| | </head> | | </head> |
| Line 29: |
Line 45: |
| | | | |
| | <div id="igem"><a href="https://2013.igem.org/Main_Page"><img class="igem" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_Uppsala_IGEM_log_blue.png"></a></div> | | <div id="igem"><a href="https://2013.igem.org/Main_Page"><img class="igem" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_Uppsala_IGEM_log_blue.png"></a></div> |
| | + | |
| | + | <img class="slogan" src="https://static.igem.org/mediawiki/2013/a/ad/Uppsala2013_Header.png"> |
| | | | |
| | </div> | | </div> |
| Line 56: |
Line 74: |
| | <li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> | | <li><a href="https://2013.igem.org/Team:Uppsala/metabolic-engineering">Metabolic engineering</a> |
| | <ul> | | <ul> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid">P-coumaric acid</a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/p-coumaric-acid">p-Coumaric acid</a></li> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/resveratrol">Resveratrol</a></li> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/lycopene">Lycopene</a></li> |
| Line 62: |
Line 80: |
| | <li><a | | <li><a |
| | href="https://2013.igem.org/Team:Uppsala/saffron">Saffron</a></li> | | href="https://2013.igem.org/Team:Uppsala/saffron">Saffron</a></li> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/astraxantin">Astaxanthin</a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/astaxanthin">Astaxanthin</a></li> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/zeaxantin">Zeaxanthin</a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/zeaxanthin">Zeaxanthin</a></li> |
| | </ul></li> | | </ul></li> |
| | </li> | | </li> |
| Line 70: |
Line 88: |
| | <li><a href="https://2013.igem.org/Team:Uppsala/chromoproteins">Chromoproteins</a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/chromoproteins">Chromoproteins</a></li> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/safety-experiment">Safety experiment</a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/safety-experiment">Safety experiment</a></li> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/result">Results</a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/results">Results</a></li> |
| | | | |
| | | | |
| Line 77: |
Line 95: |
| | <li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> | | <li><a href="https://2013.igem.org/Team:Uppsala/modeling" id="list_type1"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/6/63/Uppsala2013_Modeling.png"></a> |
| | <ul> | | <ul> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway">P-Coumaric acid</a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/P-Coumaric-acid-pathway">Kinetic model</a></li> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/modeling-tutorial">Modeling tutorial </a></li> |
| | + | |
| | + | <li><a href="https://2013.igem.org/Team:Uppsala/toxicity-model">Toxicity model</a></li> |
| | </ul></li> | | </ul></li> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/parts" id="list_type2"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/e/eb/Uppsala2013_parts.png"></a></li> |
| Line 87: |
Line 107: |
| | <li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/carotenoid-group">Carotenoid group</a></li> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/chassi-group">Chassi group</a></li> |
| | + | <li><a href="https://2013.igem.org/Team:Uppsala/advisors">Advisors</a></li> |
| | | | |
| | </ul></li> | | </ul></li> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/hp" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_HP.png"></a> | + | <li><a href="https://2013.igem.org/Team:Uppsala/human-practice" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/b/b8/Uppsala2013_HP.png"></a> |
| | <ul> | | <ul> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/Yoghurtproducts">Yoghurt +</a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/yoghurt">Yoghurt +</a></li> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/synbioday">SynBioDay</a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/synbioday">SynBioDay</a></li> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/safety">Biosafety and ethics</a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/biosafety-and-ethics">Biosafety and ethics</a></li> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/society">Public opinion </a></li> | + | <li><a href="https://2013.igem.org/Team:Uppsala/public-opinion">Public opinion </a></li> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/Outreach">High school & media </a></li> |
| - | | + | <li><a href="https://2013.igem.org/Team:Uppsala/bioart">BioArt</a></li> |
| | + | <li><a href="https://2013.igem.org/Team:Uppsala/LactonutritiousWorld">A LactoWorld</a></li> |
| | + | <li><a href="https://2013.igem.org/Team:Uppsala/killswitches">Killswitches</a></li> |
| | + | <li><a href="https://2013.igem.org/Team:Uppsala/realization">Patent</a></li> |
| | </ul></li> | | </ul></li> |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a> | + | <li><a href="https://2013.igem.org/Team:Uppsala/attribution" id="list_type4"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/5/5d/Uppsala2013_Attributions.png"></a></li> |
| - | <ul>
| + | |
| - | <li><a href="https://2013.igem.org/Team:Uppsala/collaboration">Collaboration</a></li>
| + | |
| - | </ul></li>
| + | |
| | <li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> | | <li><a href="https://2013.igem.org/Team:Uppsala/notebook" id="list_type3"><img class="nav-text" src="https://static.igem.org/mediawiki/2013/3/36/Uppsala2013_Notebook.png"></a> |
| | <ul> | | <ul> |
| | <li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> | | <li><a href="https://2013.igem.org/Team:Uppsala/safety-form">Safety form</a></li> |
| | + | <li><a href="https://2013.igem.org/Team:Uppsala/protocols">Protocols</a></li> |
| | </ul></li> | | </ul></li> |
| | </ul> | | </ul> |
| Line 113: |
Line 135: |
| | | | |
| | <div id="main_content"> <!-- Put content here --> | | <div id="main_content"> <!-- Put content here --> |
| - |
| |
| | | | |
| | + | <h1 class="main-title">Shuttle vectors for Lactobacillus and E. coli</h1> |
| | + | <div id="p-com-text1"> |
| | + | <h1>Building bridges between E. coli and Lactobacillus</h1> |
| | + | |
| | + | <p>A shuttle vector is a plasmid that can be transferred between two different species and is able to replicate in both. Such a vector is key for bioengineering in Lactobacillus. The low transformation frequencies of ligations and longer generation times slows down the work pace tremendously if you have to assembly and test everything in Lactobacillus. A solution to this problem is if you instead build everything in E. coli and then transfer the finished construct to Lactobacillus. We have created two shuttle vectors that we have sent to the registry.</p><br> |
| | + | |
| | + | |
| | + | <h3>Shuttle vectors</h3> |
| | + | |
| | + | <a href="http://parts.igem.org/Part:BBa_K1033206">BBa_K1033206, with chloramphenicol resistance.</a><br> |
| | + | <a href="http://parts.igem.org/Part:BBa_K1033207">BBa_K1033207, with erythromycin resistance.</a> |
| | + | <br><br> |
| | + | |
| | + | </div> |
| | + | <div class="text"> |
| | + | <a id="b1"></a><h1>Construction of shuttle vectors</h1> |
| | + | <p> |
| | + | To create the shuttle vector we replaced the replicon in a biobrick compatible plasmid BBa_K864003 with a replicon, pSH71, from a plasmid known to work in both E. coli and Lactobacillus. pSH71 originates from a plasmid, pJP059, from Lactococcus lactis but it is well known to replicate in both the Lactobacillus genus and in E. coli through rolling circle replication.<sup><a href="#ref_point">[1]</a></sup><br><br> |
| | + | |
| | + | Many species in the Lactobacillus genus have inherent antibiotic resistances.<sup><a href="#ref_point2">[2]</a></sup>, because of that we have been limited in our selection of antibiotic resistance. Most commonly used are chloramphenicol and erythromycin and we made versions of both for our shuttle vector. BBa_K1033207 contains an erythromycin cassette from pLUL631 and BBa_K1033206 contains the resistance cassette from pSB1C3 but has had its promoter replaced with our cp29 promoter.<sup><a href="#ref_point3">[3]</a></sup><br><br> </p> |
| | + | </div> |
| | + | <a href="https://static.igem.org/mediawiki/2013/9/92/Shuttle-vector_pSBLbC.png" data-lightbox="roadtrip"><img class="vector-plasmid" src="https://static.igem.org/mediawiki/2013/9/92/Shuttle-vector_pSBLbC.png"></a> |
| | | | |
| | + | <h1>Results</h1> |
| | + | |
| | + | <p>We have managed to transform both shuttle vectors to E. coli and verified them by sequencing. We have also managed to subclone with them and replaced the red insert with chromoproteins expressed by our CP promoters that works both in E. coli and Lactobacillus, clearly yielding blue colonies.</p> |
| | + | |
| | + | <div id="pic_type2"> |
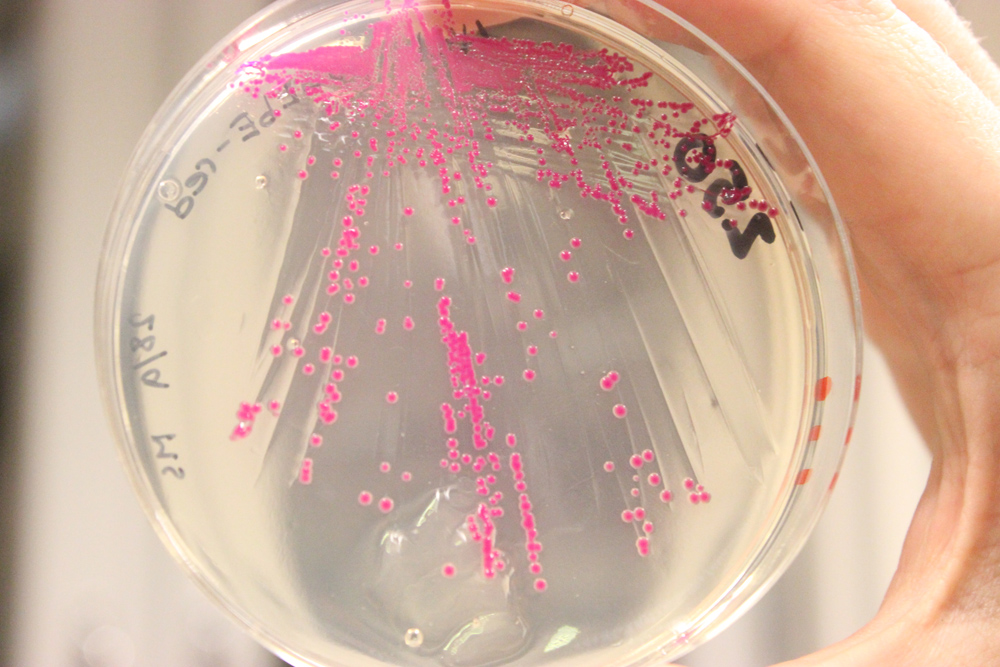
| | + | <a href="https://static.igem.org/mediawiki/2013/e/e1/IMG_6644.JPG" data-lightbox="roadtrip" title="BBa_K1033207 transformed to Lactobacillus reuteri 100-23"><img class="pic_type2" src="https://static.igem.org/mediawiki/2013/e/e1/IMG_6644.JPG"></a> |
| | + | |
| | + | <i>To the left, BBa_K1033207 transformed to Lactobacillus reuteri 100-23, to the right, negative control without plasmid</i> |
| | + | </div> |
| | + | |
| | + | <div id="pic_type3"> |
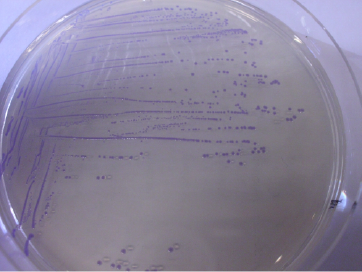
| | + | <a href="https://static.igem.org/mediawiki/2013/2/28/Uppsala2013_chromo.JPG" data-lightbox="roadtrip" title="BBa_K1033207 and RFP, BBa_J04450 insert in E. coli"><img class="pic_type3" src="https://static.igem.org/mediawiki/2013/2/28/Uppsala2013_chromo.JPG"></a> |
| | + | |
| | + | <i class="middle-text">BBa_K1033207 and RFP, BBa_J04450 |
| | + | <br> |
| | + | <i class="middle-text">insert in E. coli</i> |
| | + | </div> |
| | + | |
| | + | <div id="pic_type4"> |
| | + | <a href="https://static.igem.org/mediawiki/2013/b/ba/Uppsala2013_Shuttle_Vector_pSBLBC_cp29_amilCP.JPG" data-lightbox="roadtrip" title="BBa_K1033206 with a subcloned chromoprotein BBa_K1033282 in E. coli"><img class="shuttle_vec" src="https://static.igem.org/mediawiki/2013/1/18/Uppsala2013_Shuttle_Vector_pSBLBC_cp29_amilCP1.png"></a> |
| | + | |
| | + | <i>BBa_K1033206 with a subcloned chromoprotein BBa_K1033282 in E. coli</i> |
| | + | </div> |
| | + | |
| | + | <div id="clear"></div> |
| | + | |
| | + | <p>We have also gained positive results when transforming BBa_K1033207 to Lactobacillus reuteri and Lactobacillus plantarum. <p> |
| | + | |
| | + | <div id="reference"> |
| | + | <h1>Reference</h1> |
| | + | <p class="reference"> |
| | + | |
| | + | <a id="ref_point">[1]</a> <a href="http://www.ncbi.nlm.nih.gov/pubmed/11591136"> Construction of compatible wide-host-range shuttle v... </a> [Plasmid. 2001] - PubMed - NCBI." National Center for Biotechnology Information. N.p., n.d. Web. 1 Oct. 2013. |
| | + | |
| | + | <br><br> |
| | + | |
| | + | <a id="ref_point2">[2]</a> <a href="http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3378904"> "High efficiency recombineering in lactic acid bacteria. </a> National Center for Biotechnology Information. N.p., n.d. Web. 1 Oct. 2013. |
| | + | <br><br> |
| | + | |
| | + | <a id="ref_point3">[3]</a> <a href="http://www.researchgate.net/publication/226939823_Transformation_ofLactobacillus_reuteri_with_electroporation_Studies_on_the_erythromycin_resistance_plasmid_pLUL631?ev=pub_cit%22,%22http://www.researchgate.net/publication/226939823_Transformation_ofLactobacillus_reuteri_with_electroporation_Studies_on_the_erythromycin_resistance_plasmid_pLUL631?ev=pub_cit%22) |
| | + | "> Transformation of Lactobacillus reuteri with electroporation: Studies on the erythromycin resistance plasmid pLUL631 </a> , Siv Ahrné, Göran Molin, Lars Axelsson, Januari 1992 |
| | + | |
| | + | <br><br> |
| | + | |
| | + | </p> |
| | + | |
| | + | </div> |
| | | | |
| | </div> <!-- main_ontent ends --> | | </div> <!-- main_ontent ends --> |
| | | | |
| | <div id="bottom-pic"> | | <div id="bottom-pic"> |
| - | <img class="bottom-pic" src="https://static.igem.org/mediawiki/2013/a/aa/Bottom_picture.png"> | + | |
| | </div> | | </div> |
| | | | |
 "
"