Team:Evry/Protocols/11
From 2013.igem.org
(Created page with "{{:Team:Evry/template_protocols}} <html> <div id="mainTextcontainer"> <!--<a href='https://2013.igem.org/Team:Evry/Protocoles/08' title='Vers la page française'> <img src='htt...") |
|||
| (19 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<h1> PCR Purification </h1> | <h1> PCR Purification </h1> | ||
| - | <h2> | + | <h2> Goal </h2> |
| - | <p></p> | + | <p>The aim of the PCR purification step is to recover DNA from <a href="https://2013.igem.org/Team:Evry/Protocols/07" target='_blank'>PCR</a> and other enzymatic reaction mixtures.</p> |
<h2> Preparation </h2> | <h2> Preparation </h2> | ||
| Line 17: | Line 17: | ||
| - | <b><p> | + | <b><p>1. </b><br> |
| + | Add 1:1 volume of Binding Buffer to completed PCR mixture (e.g. if you have 100 µL of reaction mixture, add 100 µL of Binding Buffer). | ||
| + | Mix thoroughly and check the solution's color: a yellow color color indicates an optimal pH for DNA binding. | ||
| + | <br>If the color of the solution is orange or violet, add 10 µL of 3 M sodium acetate, pH 5,2 and mix. The color will become yellow.<br> | ||
| + | If the DNA fragment is superior to 500 bp, add a 1:2 volume of 100% isopropanol (e.g. if you have 100 µL of PCR mixture combined with 100 µL <i>(total amount of 200 µL)</i>, add 100 µL isopropanol). Mix thoroughly.</p> | ||
| + | <br> | ||
| + | <p><b>2. </b><br>Transfer up 800 µL of the solution from step previous step to the GeneJET purification column.<br> | ||
| + | Centrifuge for 30-60 seconds, then discard the flow-through. | ||
| + | </p> | ||
| + | <br> | ||
| + | |||
| + | <p><b>3. </b> <br> | ||
| + | Add 700 µL of Wash Buffer (previously dilted with ethanol) to the purification column. <br> | ||
| + | Centrifuge for 30-60 seconds, then discard the flow-through and place the purification column back into the collection tube? | ||
| + | </p> | ||
| + | <br> | ||
| + | |||
| + | <p><b>4. </b><br> | ||
| + | Centrifuge the empty GeneJET purification for 1 minute to completely remove any residual wash buffer<br><br></p> | ||
| + | |||
| + | <p><b>5. <br></b> | ||
| + | Transfer the GeneJET purification column to a clean 1,5 mL tube.<br> | ||
| + | Add 50 µL of Elution Buffer to the center of the GeneJET purification column membrane and centrifuge for 1 minute.</p> | ||
| + | <br> | ||
| + | |||
| + | <p><b>6. <br></b> | ||
| + | Discard the GeneJET purification column and store the purified DNA at -20°C.</p> | ||
| + | <br><br> | ||
| + | |||
| + | |||
| + | <h2> Test </h2> | ||
| + | |||
| + | <div class="center"> | ||
| + | <div class="thumb tnone"> | ||
| + | <div class="thumbinner" style="width:502px;"> | ||
| + | <a href="https://static.igem.org/mediawiki/2013/f/f0/Nanodrop.png" class="image"> | ||
| + | <img alt="" src="https://static.igem.org/mediawiki/2013/f/f0/Nanodrop.png" width="500" class="thumbimage" /></a> <div class="thumbcaption"> | ||
| + | <div class="magnify"> | ||
| + | <a href="https://static.igem.org/mediawiki/2013/f/f0/Nanodrop.png" class="internal" title="Enlarge"> | ||
| + | <img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a> | ||
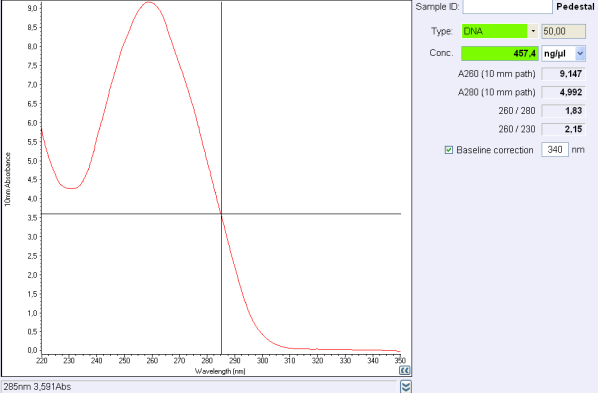
| + | </div >Figure 1: Nanodrop test after a plasmid purification. | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | If the concentration is between .. and .. ng/μL, concentrate your DNA with SpeedVac concentrator or any other method.<br> | ||
| + | If the concentration is below .. ng/μL or if 260/280 ratio or 260/230 ratio is not correct, make another purification.<br><br> | ||
| + | |||
| + | <i>260/280 ratio and 260/230 indicate the purity of DNA (or RNA). <br> | ||
| + | Nucleic acids absorb at 260 nm while proteins and phenols absorb at 280 nm and carbohydrates at 230 and other contaminents at 230 nm.<br> | ||
| + | For DNA, a 260/280 ratio around 1,8 is concider to be pure and ange 260/230 ratio must be between 2,0 and 2,2.</i><br></p> | ||
</div> | </div> | ||
Latest revision as of 09:55, 2 September 2013
PCR Purification
Goal
The aim of the PCR purification step is to recover DNA from PCR and other enzymatic reaction mixtures.
Preparation
Protocol adapted from Thermo Scientific PCR Purification notebook1.
Add 1:1 volume of Binding Buffer to completed PCR mixture (e.g. if you have 100 µL of reaction mixture, add 100 µL of Binding Buffer).
Mix thoroughly and check the solution's color: a yellow color color indicates an optimal pH for DNA binding.
If the color of the solution is orange or violet, add 10 µL of 3 M sodium acetate, pH 5,2 and mix. The color will become yellow.
If the DNA fragment is superior to 500 bp, add a 1:2 volume of 100% isopropanol (e.g. if you have 100 µL of PCR mixture combined with 100 µL (total amount of 200 µL), add 100 µL isopropanol). Mix thoroughly.
2.
Transfer up 800 µL of the solution from step previous step to the GeneJET purification column.
Centrifuge for 30-60 seconds, then discard the flow-through.
3.
Add 700 µL of Wash Buffer (previously dilted with ethanol) to the purification column.
Centrifuge for 30-60 seconds, then discard the flow-through and place the purification column back into the collection tube?
4.
Centrifuge the empty GeneJET purification for 1 minute to completely remove any residual wash buffer
5.
Transfer the GeneJET purification column to a clean 1,5 mL tube.
Add 50 µL of Elution Buffer to the center of the GeneJET purification column membrane and centrifuge for 1 minute.
6.
Discard the GeneJET purification column and store the purified DNA at -20°C.
Test
If the concentration is between .. and .. ng/μL, concentrate your DNA with SpeedVac concentrator or any other method.If the concentration is below .. ng/μL or if 260/280 ratio or 260/230 ratio is not correct, make another purification.
260/280 ratio and 260/230 indicate the purity of DNA (or RNA).
Nucleic acids absorb at 260 nm while proteins and phenols absorb at 280 nm and carbohydrates at 230 and other contaminents at 230 nm.
For DNA, a 260/280 ratio around 1,8 is concider to be pure and ange 260/230 ratio must be between 2,0 and 2,2.
 "
"