Team:Paris Saclay/Notebook/August/9
From 2013.igem.org
(Difference between revisions)
(→1 - Tranduction of Km in MG1655Z1) |
|||
| (16 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
==='''A - Aerobic/Anaerobic regulation system'''=== | ==='''A - Aerobic/Anaerobic regulation system'''=== | ||
| - | + | ===='''Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006'''==== | |
| - | + | ===='''1 - Tranduction of Km in MG1655Z1 ==== | |
| - | + | Abdou, Anaïs, Damir, Nadia, XiaoJing | |
| - | + | {| | |
| + | | style="border:1px solid black;padding:5px;background-color:#DE;" | | ||
| + | We observed lysis areas in the strain MG1655Z1 Δfnr::Km after transduction. We will continue the transduction protocol. | ||
| + | |} | ||
{| | {| | ||
| Line 21: | Line 24: | ||
* 0µl phage(control): the petri dish is cloudy, bacteria are not lysed. | * 0µl phage(control): the petri dish is cloudy, bacteria are not lysed. | ||
* 50µl phage: the petri dish is clear, bacteria are lysed by phages. | * 50µl phage: the petri dish is clear, bacteria are lysed by phages. | ||
| - | |||
|} | |} | ||
| - | + | ===='''Objective : obtaining BBa_K1155007'''==== | |
| - | + | ||
| - | + | ||
| - | + | ====1 - Extraction of BBa_K115007 from DH5αstrain==== | |
| - | + | ||
| - | + | ||
| - | ====1 - Extraction of | + | |
Abdou | Abdou | ||
| - | Protocol : [[Team:Paris_Saclay/ | + | Protocol : [[Team:Paris_Saclay/extraction|High-copy plamid extraction]] |
| - | We | + | We extracted plamid from colonies number 10, 14 and 15. |
Nanodrop | Nanodrop | ||
| - | * | + | * BBa_K1155007 in clone 10 : 38ng/µl |
| - | * | + | * BBa_K1155007 in clone 14 : 48.5ng/µl |
| - | * | + | * BBa_K1155007 in clone 15 : 52 ng/µl |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | + | The extraction was good. We will sequence our plasmids. | |
|} | |} | ||
| - | |||
==='''A - Aerobic/Anaerobic regulation system / B - PCB sensing system'''=== | ==='''A - Aerobic/Anaerobic regulation system / B - PCB sensing system'''=== | ||
| - | Objective : Obtaining FNR and BphR2 proteins | + | ====Objective : Obtaining FNR and BphR2 proteins==== |
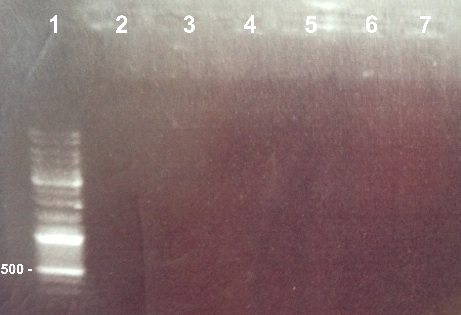
| - | + | ===='''1 - Electrophoresis of the PCR of BphR2 Part I, BphR2 Part II, RBS_BphR2 Part I, FNR Part I, FNR Part II, RBS_FNR Part I to check the gel purification'''==== | |
| - | + | ||
| - | BphR2 | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| | {| | ||
| - | | style="width:350px;border:1px solid black;" | | + | | style="width:350px;border:1px solid black;" |[[File:Psgel10908.jpg]] |
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
*Well 1 : 6µL DNA Ladder | *Well 1 : 6µL DNA Ladder | ||
| - | *Well 2 : 5µL of BphR2 | + | *Well 2 : 5µL of BphR2 Part I + 1µl of 6X loading dye |
| - | *Well 3 : 5µL of BphR2 | + | *Well 3 : 5µL of BphR2 Part II + 1µl of 6X loading dye |
| - | *Well 4 : 5µL of | + | *Well 4 : 5µL of RBS-BphR2 Part I + 1µl of 6X loading dye |
| - | *Well 5 : 5µL of FNR | + | *Well 5 : 5µL of FNR Part I + 1µl of 6X loading dye |
| - | *Well 6 : 5µL of FRN | + | *Well 6 : 5µL of FRN Part II + 1µl of 6X loading dye |
| - | *Well 7: 5µL of | + | *Well 7: 5µL of RBS-FNR Part I + 1µl of 6X loading dye |
*Gel : 0.8% | *Gel : 0.8% | ||
|} | |} | ||
| - | + | Expected size : | |
| + | * BphR2 Part I : 178 bp | ||
| + | * BphR2 Part II : 790bp | ||
| + | * RBS-BphR2 Part I : 197bp | ||
| + | * FNR Part I : 597 bp | ||
| + | * FNR Part II : 200bp | ||
| + | * RBS-FNR PartI : 615bp | ||
| + | |||
| + | {| | ||
| + | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| + | We lost all our PCR fragments. We will do the PCR again. | ||
| + | |} | ||
| + | |||
| + | ===='''2 - PCR of BphR2 Part I, BphR2 Part II, RBS-BphR2 Part I, FNR Part I, FNR Part II, RBS-FNR Part I'''==== | ||
| + | |||
| + | Anaïs, Damir, Nadia, XiaoJing | ||
| + | |||
| + | Used quantities : | ||
| + | * Bphr2 Part I : | ||
| + | ** Oligo 54F : 1µL | ||
| + | ** Oligo 55R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA of ''Pseudomonas pseudoalcaligenes'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | * Bphr2 Part II : | ||
| + | ** Oligo 56F : 1µL | ||
| + | ** Oligo 57R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA ''Pseudomonas pseudoalcaligenes'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | * RBS-Bphr2 Part I : | ||
| + | ** Oligo 58F : 1µL | ||
| + | ** Oligo 57R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA ''Pseudomonas pseudoalcaligenes'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | * FNR Part I : | ||
| + | ** Oligo 59F : 1µL | ||
| + | ** Oligo 60R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA ''Escherichia coli'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | * FNR Part II : | ||
| + | ** Oligo 61F : 1µL | ||
| + | ** Oligo 62R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA ''Escherichia coli'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | * RBS-FNR Part I : | ||
| + | ** Oligo 63F : 1µL | ||
| + | ** Oligo 62R : 1µL | ||
| + | ** Buffer Phusion : 10µL | ||
| + | ** DNA ''Escherichia coli'' : 1µL | ||
| + | ** dNTP : 1µL | ||
| + | ** Phusion : 0.5µL | ||
| + | ** H2O : 35.5µL | ||
| + | |||
| + | PCR Program : | ||
| + | |||
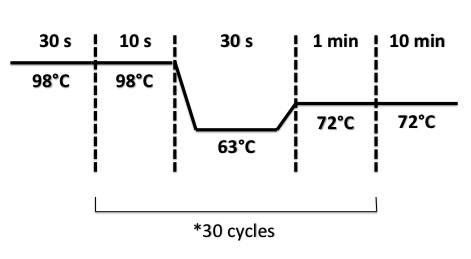
| + | * BphR2 Part I, BphR2 Part II, RBS-BphR2 Part I : | ||
| + | |||
| + | [[File:PsPCRBphR23007.jpg|400px]] | ||
| + | |||
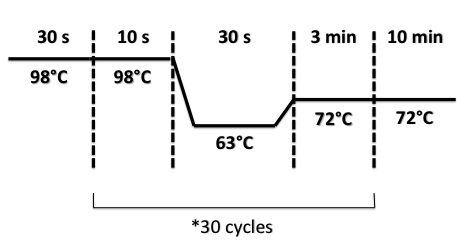
| + | * FNR Part I, FNR Part II, RBS-FNR Part I : | ||
| + | |||
| + | [[File:PsPCRFNR3007.jpg|400px]] | ||
| + | |||
| + | |||
| + | {| border="1" align="center" | ||
| + | |[[Team:Paris Saclay/Notebook/August/8|<big>Previous day</big>]] | ||
| + | |||
| + | |[[Team:Paris_Saclay/Notebook|<big>Back to calendar</big>]] | ||
| + | |||
| + | |[[Team:Paris Saclay/Notebook/August/12|<big>Next day</big>]] | ||
| + | |} | ||
| + | |||
{{Team:Paris_Saclay/incl_fin}} | {{Team:Paris_Saclay/incl_fin}} | ||
Latest revision as of 15:28, 4 October 2013
Notebook : August 9
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006
1 - Tranduction of Km in MG1655Z1
Abdou, Anaïs, Damir, Nadia, XiaoJing
|
We observed lysis areas in the strain MG1655Z1 Δfnr::Km after transduction. We will continue the transduction protocol. |
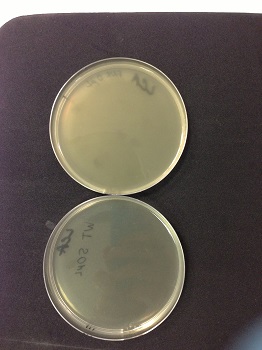
|
Picture: lysed cells comparison.
|
Objective : obtaining BBa_K1155007
1 - Extraction of BBa_K115007 from DH5αstrain
Abdou
Protocol : High-copy plamid extraction
We extracted plamid from colonies number 10, 14 and 15.
Nanodrop
- BBa_K1155007 in clone 10 : 38ng/µl
- BBa_K1155007 in clone 14 : 48.5ng/µl
- BBa_K1155007 in clone 15 : 52 ng/µl
|
The extraction was good. We will sequence our plasmids. |
A - Aerobic/Anaerobic regulation system / B - PCB sensing system
Objective : Obtaining FNR and BphR2 proteins
1 - Electrophoresis of the PCR of BphR2 Part I, BphR2 Part II, RBS_BphR2 Part I, FNR Part I, FNR Part II, RBS_FNR Part I to check the gel purification
Expected size :
- BphR2 Part I : 178 bp
- BphR2 Part II : 790bp
- RBS-BphR2 Part I : 197bp
- FNR Part I : 597 bp
- FNR Part II : 200bp
- RBS-FNR PartI : 615bp
|
We lost all our PCR fragments. We will do the PCR again. |
2 - PCR of BphR2 Part I, BphR2 Part II, RBS-BphR2 Part I, FNR Part I, FNR Part II, RBS-FNR Part I
Anaïs, Damir, Nadia, XiaoJing
Used quantities :
- Bphr2 Part I :
- Oligo 54F : 1µL
- Oligo 55R : 1µL
- Buffer Phusion : 10µL
- DNA of Pseudomonas pseudoalcaligenes : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- Bphr2 Part II :
- Oligo 56F : 1µL
- Oligo 57R : 1µL
- Buffer Phusion : 10µL
- DNA Pseudomonas pseudoalcaligenes : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- RBS-Bphr2 Part I :
- Oligo 58F : 1µL
- Oligo 57R : 1µL
- Buffer Phusion : 10µL
- DNA Pseudomonas pseudoalcaligenes : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- FNR Part I :
- Oligo 59F : 1µL
- Oligo 60R : 1µL
- Buffer Phusion : 10µL
- DNA Escherichia coli : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- FNR Part II :
- Oligo 61F : 1µL
- Oligo 62R : 1µL
- Buffer Phusion : 10µL
- DNA Escherichia coli : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
- RBS-FNR Part I :
- Oligo 63F : 1µL
- Oligo 62R : 1µL
- Buffer Phusion : 10µL
- DNA Escherichia coli : 1µL
- dNTP : 1µL
- Phusion : 0.5µL
- H2O : 35.5µL
PCR Program :
- BphR2 Part I, BphR2 Part II, RBS-BphR2 Part I :
- FNR Part I, FNR Part II, RBS-FNR Part I :
| Previous day | Back to calendar | Next day |
 "
"