Team:Bielefeld-Germany/Labjournal/May
From 2013.igem.org
(Difference between revisions)
| (17 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
__NOTOC__ | __NOTOC__ | ||
| + | <div id=globalwrapper style="padding-left:20px; padding-right:20px"> | ||
| + | <div id="leftcol" style="width:750px; float:left; overflow:auto;"> | ||
<html> | <html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<style> | <style> | ||
| - | h1{ | + | h1{margin-top:20px;} |
| - | + | ||
</style> | </style> | ||
| - | |||
| - | |||
| - | |||
| - | |||
<h1>May</h1> | <h1>May</h1> | ||
</html> | </html> | ||
| Line 30: | Line 19: | ||
| - | *Initiating | + | *Initiating lab work on the sub-project endogenous mediator [[Team:Bielefeld-Germany/Project/GldA|Glycerol dehydrogenase]]. |
| - | *Starting lab work with the first successful PCR: | + | *Starting lab work with the first successful PCR: ''gldA'' with pre- and suffix overlaps could be amplified from ''Escherichia coli'' genome. |
| - | *The main work however is still planning of our | + | *The main work however is still planning of our sub-projects, designing experiments and a lot of research. |
*Finding sponsors goes ahead. Many companies like our project and want to support us. | *Finding sponsors goes ahead. Many companies like our project and want to support us. | ||
| - | + | *MFC: We built a first prototype of a microbial fuel cell and tested it with different chemicals, baker’s yeast and the ''E. coli'' KRX wild type strain. Tested if methylene blue and neutral red are suitable as redox mediators for our system. | |
| + | <br><br> | ||
| Line 45: | Line 35: | ||
====MFC==== | ====MFC==== | ||
| - | + | *Borrowed 2 multimeters from the faculty of physics. | |
====Mediators==== | ====Mediators==== | ||
*Glycerol dehydrogenase | *Glycerol dehydrogenase | ||
| - | **Primerdesign for isolation of | + | **Primerdesign for isolation of ''gldA'' gene from ''Escherichia coli'' KRX strain, with overlaps for BioBrick prefix and suffix: |
| - | **Forward | + | **Forward primer ''gldA'' (43 bp): GAATTCGCGGCCGCTTCTAGATGGACCGCATTATTCAATCACC |
| - | **Reverse | + | **Reverse primer ''gldA'' (45 bp): CTGCAGCGGCCGCTACTAGTATTATTCCCACTCTTGCAGGAAACG |
| - | + | <br><br> | |
| - | + | ||
| - | === | + | ===Week 2=== |
====Organization==== | ====Organization==== | ||
*Distribution of our team in various subgroups for best work efficiency. | *Distribution of our team in various subgroups for best work efficiency. | ||
| - | *We’ve organized a | + | *We’ve organized a barbecue to thank the working group of Dr. Jörn Kalinowski in the CeBiTec for the appropriation of space and support in the laboratory. |
====MFC==== | ====MFC==== | ||
| + | *Constructed a first prototype of a fuel cell out of two film canisters connected with a fragment of a centrifugation tube. | ||
| + | <br><br> | ||
| - | |||
| - | |||
| - | === | + | ===Week 3=== |
====Organization==== | ====Organization==== | ||
| - | *Planning the trip to the European jamboree in Lyon from | + | *Planning the trip to the European jamboree in Lyon from October 11th to 13th, 2013. |
====MFC==== | ====MFC==== | ||
| - | + | *Experimented with with the film canister fuel cell using yeast as a biocatalyst. | |
====Mediators==== | ====Mediators==== | ||
| Line 85: | Line 75: | ||
**Starting first cultivation of ''Escherichia coli'' KRX strain for [[Team:Bielefeld-Germany/Labjournal/Molecular#Whole Genome Isolation |complete genome isolation]]. | **Starting first cultivation of ''Escherichia coli'' KRX strain for [[Team:Bielefeld-Germany/Labjournal/Molecular#Whole Genome Isolation |complete genome isolation]]. | ||
**Successful [[Team:Bielefeld-Germany/Labjournal/Molecular#Whole Genome Isolation| genome isolation]] of ''Escherichia coli'' KRX. | **Successful [[Team:Bielefeld-Germany/Labjournal/Molecular#Whole Genome Isolation| genome isolation]] of ''Escherichia coli'' KRX. | ||
| - | **Successful PCR on the | + | **Successful PCR on the ''gldA'' gene of ''Escherichia coli'' KRX strain. |
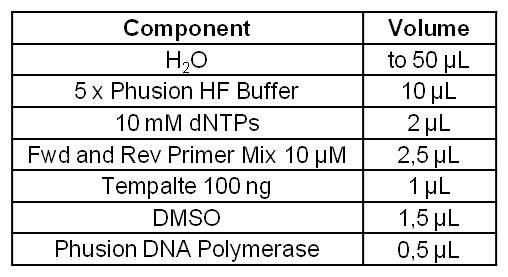
| - | [[Image:IGEM_Bielefeld_Standard_Phusion_PCR_Master_MixLRO.jpg|300px|thumb|left| <p align="justify">'''Table 1: Standard | + | [[Image:IGEM_Bielefeld_Standard_Phusion_PCR_Master_MixLRO.jpg|300px|thumb|left| <p align="justify">'''Table 1: Standard phusion PCR master mix. '''</p>]] |
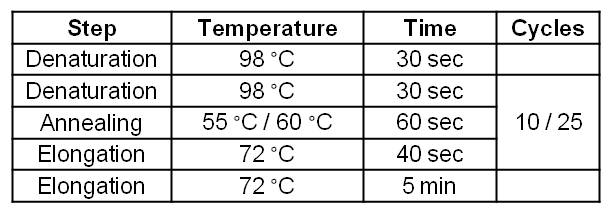
| - | [[Image:IGEM_Bielefeld_Standard_Phu_PCR_GldA_OprF.jpg|300px|thumb|center| <p align="justify">'''Table 2: Two step standard | + | [[Image:IGEM_Bielefeld_Standard_Phu_PCR_GldA_OprF.jpg|300px|thumb|center| <p align="justify">'''Table 2: Two step standard phusion PCR program for ''gldA'' amplification. '''</p>]] |
| - | ** | + | **The ''gldA'' PCR product was isolated by agarose gel electrophoresis and [[Team:Bielefeld-Germany/Labjournal/Molecular#Used Kits| purified]]. |
| - | **Bands are at expected size of 1050 bp. | + | **Bands are on at the expected size of 1050 bp. |
| - | [[Image:IGEM_Bielefeld_GldA_PCR_Gel.jpg|200px|thumb|left| <p align="justify">'''Figure 1: | + | [[Image:IGEM_Bielefeld_GldA_PCR_Gel.jpg|200px|thumb|left| <p align="justify">'''Figure 1: Agarose gel from PCR on the ''gldA'' gene of ''Escherichia coli'' KRX strain with forward and reverse primer ''gldA''. As a ladder we used [http://www.thermoscientificbio.com/nucleic-acid-electrophoresis/generuler-1-kb-dna-ladder-ready-to-use-250-to-10000-bp GeneRuler™ 1 kb DNA Ladder from Thermo Scientific]. '''</p>]] |
| - | + | <br><br> | |
| - | + | ||
| - | === | + | ===Week 4=== |
====Organization==== | ====Organization==== | ||
| - | *We will also take part at the congress | + | *We will also take part at the congress ‘‘Next generation of biotechnological processes 2020+‘‘ in Berlin on June 27th, 2013. |
*Having a second presentation at Merck KGaA head office in Darmstadt for explaining our project in detail. We are happy that Merck will be our next supporter in 2013. | *Having a second presentation at Merck KGaA head office in Darmstadt for explaining our project in detail. We are happy that Merck will be our next supporter in 2013. | ||
| - | + | ===MFC=== | |
| - | + | *Tested if methylene blue and neutral red can work as redox mediators in our system. | |
| - | + | ||
<br><br><br><br> | <br><br><br><br> | ||
| Line 117: | Line 106: | ||
| - | <div id="asdf" | + | <div id="asdf" class="withMenu"> |
<html> | <html> | ||
<div id="nav2" style="width:210px; padding-bottom:5px; padding-left:15px;"> | <div id="nav2" style="width:210px; padding-bottom:5px; padding-left:15px;"> | ||
| Line 145: | Line 134: | ||
| - | <div id="rightcol" style="width:210px; height: | + | <div id="rightcol" style="width:210px; height:100%; overflow-y:auto; box-shadow:0px 0px 2px 0px grey;" padding:0px 20px;> |
__TOC__ | __TOC__ | ||
<div id="spacer" style="height:300px"></div> | <div id="spacer" style="height:300px"></div> | ||
Latest revision as of 23:56, 28 October 2013
May
Milestones
- Initiating lab work on the sub-project endogenous mediator Glycerol dehydrogenase.
- Starting lab work with the first successful PCR: gldA with pre- and suffix overlaps could be amplified from Escherichia coli genome.
- The main work however is still planning of our sub-projects, designing experiments and a lot of research.
- Finding sponsors goes ahead. Many companies like our project and want to support us.
- MFC: We built a first prototype of a microbial fuel cell and tested it with different chemicals, baker’s yeast and the E. coli KRX wild type strain. Tested if methylene blue and neutral red are suitable as redox mediators for our system.
Week 1
Organization
- We were already able to find sponsors for our team: IIT BIEKUBA, IIT Biotech, Evonik, PlasmidFactory and FisherScientific will support us.
MFC
- Borrowed 2 multimeters from the faculty of physics.
Mediators
- Glycerol dehydrogenase
- Primerdesign for isolation of gldA gene from Escherichia coli KRX strain, with overlaps for BioBrick prefix and suffix:
- Forward primer gldA (43 bp): GAATTCGCGGCCGCTTCTAGATGGACCGCATTATTCAATCACC
- Reverse primer gldA (45 bp): CTGCAGCGGCCGCTACTAGTATTATTCCCACTCTTGCAGGAAACG
Week 2
Organization
- Distribution of our team in various subgroups for best work efficiency.
- We’ve organized a barbecue to thank the working group of Dr. Jörn Kalinowski in the CeBiTec for the appropriation of space and support in the laboratory.
MFC
- Constructed a first prototype of a fuel cell out of two film canisters connected with a fragment of a centrifugation tube.
Week 3
Organization
- Planning the trip to the European jamboree in Lyon from October 11th to 13th, 2013.
MFC
- Experimented with with the film canister fuel cell using yeast as a biocatalyst.
Mediators
- Glycerol dehydrogenase
- Starting first cultivation of Escherichia coli KRX strain for complete genome isolation.
- Successful genome isolation of Escherichia coli KRX.
- Successful PCR on the gldA gene of Escherichia coli KRX strain.
- The gldA PCR product was isolated by agarose gel electrophoresis and purified.
- Bands are on at the expected size of 1050 bp.

Figure 1: Agarose gel from PCR on the gldA gene of Escherichia coli KRX strain with forward and reverse primer gldA. As a ladder we used [http://www.thermoscientificbio.com/nucleic-acid-electrophoresis/generuler-1-kb-dna-ladder-ready-to-use-250-to-10000-bp GeneRuler™ 1 kb DNA Ladder from Thermo Scientific].
Week 4
Organization
- We will also take part at the congress ‘‘Next generation of biotechnological processes 2020+‘‘ in Berlin on June 27th, 2013.
- Having a second presentation at Merck KGaA head office in Darmstadt for explaining our project in detail. We are happy that Merck will be our next supporter in 2013.
MFC
- Tested if methylene blue and neutral red can work as redox mediators in our system.
 "
"