Team:Paris Saclay/Notebook/August/14
From 2013.igem.org
(→2 - Electrophoresis of PCR products : BphR2 Part I) |
(→9 - Gel purification of the digestion of BBa_K1155003 and BBa_K1155007 by XBaI/PstI) |
||
| (10 intermediate revisions not shown) | |||
| Line 35: | Line 35: | ||
* NarG : 80.7ng/µL | * NarG : 80.7ng/µL | ||
* Nir B :65.8ng/µL | * Nir B :65.8ng/µL | ||
| - | * | + | * Pndh* : 227ng/µL |
{| | {| | ||
| Line 102: | Line 102: | ||
| style="width:350px;border:1px solid black;" | [[File:PSgel3_1408.jpg]] | | style="width:350px;border:1px solid black;" | [[File:PSgel3_1408.jpg]] | ||
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
| - | *Well 1 : 5µL of BBa_K1155000 digested by SpeI +1µl of 6X loading dye | + | *Well 1 : 5µL of BBa_K1155000 digested by SpeI + 1µl of 6X loading dye |
| - | *Well 2 : 5µL of BBa_K1155006 digested by SpeI +1µl of 6X loading dye | + | *Well 2 : 5µL of BBa_K1155006 digested by SpeI + 1µl of 6X loading dye |
| - | *Well 3 : 5µL of BBa_K1155005 digested by SpeI +1µl of 6X loading dye | + | *Well 3 : 5µL of BBa_K1155005 digested by SpeI + 1µl of 6X loading dye |
| - | *Well 4 : 5µL of BBa_K1155004 digested by SpeI +1µl of 6X loading dye | + | *Well 4 : 5µL of BBa_K1155004 digested by SpeI + 1µl of 6X loading dye |
*Well 5 : 6µL DNA Ladder | *Well 5 : 6µL DNA Ladder | ||
*Well 6 : 5µL of BBa_K1155000 digested by EcoRI/Spe I + 1µl of 6X loading dye | *Well 6 : 5µL of BBa_K1155000 digested by EcoRI/Spe I + 1µl of 6X loading dye | ||
| Line 116: | Line 116: | ||
Expected sizes : | Expected sizes : | ||
| - | * | + | *Pndh* in pSB1C3 : 2200bp |
*Nar K in pSB1C3, NarG in pSB1C3, NirB in pSB1C3 : 2270bp | *Nar K in pSB1C3, NarG in pSB1C3, NirB in pSB1C3 : 2270bp | ||
*pSB1C3 : 2070kb | *pSB1C3 : 2070kb | ||
*NarK, NarG, NirB : 200kb | *NarK, NarG, NirB : 200kb | ||
| - | * | + | *Pndh* : 111bp |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
Well 1 : we have done one digestion so we have to obtain one fragment, we have two stripes, the digestion of BBa_K1155000 by SpeI wasn't good. | Well 1 : we have done one digestion so we have to obtain one fragment, we have two stripes, the digestion of BBa_K1155000 by SpeI wasn't good. | ||
| - | Well 2, 3, 4, 6, 7 : we obtain NarG, NarK and NirB in | + | Well 2, 3, 4, 6, 7 : we obtain NarG, NarK and NirB in pSB1C3 and Pndh*, NarK fragments at the right size, we can purify it. |
Well 8, 9 : we have done two digestions so we have to obtain two fragments, we have only one stripe, the digestion of BBa_K1155005 and BBa_K1155004 by EcoRI/SpeI wasn't good. | Well 8, 9 : we have done two digestions so we have to obtain two fragments, we have only one stripe, the digestion of BBa_K1155005 and BBa_K1155004 by EcoRI/SpeI wasn't good. | ||
|} | |} | ||
| Line 142: | Line 142: | ||
Nanodrop : | Nanodrop : | ||
| - | * | + | * Pndh* : 13.7ng/µL |
* NarK : 27.9ng/µL | * NarK : 27.9ng/µL | ||
* NarK in PSB1C3 : 87.1ng/µL | * NarK in PSB1C3 : 87.1ng/µL | ||
| Line 169: | Line 169: | ||
Expected sizes : | Expected sizes : | ||
* RBS_LacZ-Term : 3500bp | * RBS_LacZ-Term : 3500bp | ||
| - | * RBS_AmilCP-Term : | + | * RBS_AmilCP-Term : 824bp |
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We | + | We obtained fragments at the right size. We will purify it. |
|} | |} | ||
| Line 185: | Line 185: | ||
{| | {| | ||
| style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | | style="border:1px solid black;padding:5px;background-color:#DEDEDE;" | | ||
| - | We | + | We obtained low quantities of plasmid that's why we will make an Ethanol precipitation to concentrate it. |
|} | |} | ||
| Line 193: | Line 193: | ||
Used quantities : | Used quantities : | ||
| - | * Glycerol : 500 | + | * Glycerol : 500 µL |
| - | * | + | * MG1655Z1 Δfnr::Km: 1 mL |
| - | + | ||
| - | + | ||
| + | We stored the bacteria at -20°C. | ||
==='''A - Aerobic/Anaerobic regulation system / B - PCB sensor system'''=== | ==='''A - Aerobic/Anaerobic regulation system / B - PCB sensor system'''=== | ||
| Line 208: | Line 207: | ||
* RBS-BphR2 : | * RBS-BphR2 : | ||
| - | ** | + | ** pSB1C3 : 3µL |
** BphR2 Part I : 1µL | ** BphR2 Part I : 1µL | ||
** BphR2 Part II : 1µL | ** BphR2 Part II : 1µL | ||
| Line 214: | Line 213: | ||
* FNR : | * FNR : | ||
| - | ** | + | ** pSB1C3 : 3µL |
** FNR Part I : 1µL | ** FNR Part I : 1µL | ||
** FNR Part II : 1µL | ** FNR Part II : 1µL | ||
| Line 220: | Line 219: | ||
* RBS-FNR : | * RBS-FNR : | ||
| - | ** | + | ** pSB1C3 : 3µL |
** RBS-FNR Part I : 1µL | ** RBS-FNR Part I : 1µL | ||
** FNR Part II : 1µL | ** FNR Part II : 1µL | ||
** Gibson mix : 15µL | ** Gibson mix : 15µL | ||
| - | We incubate these mix at 50°C during 1h. Then we | + | We incubate these mix at 50°C during 1h inside PCR machine. Then we keep these mix at 4°C during the week end. |
===='''2 - Electrophoresis of PCR products : BphR2 Part I'''==== | ===='''2 - Electrophoresis of PCR products : BphR2 Part I'''==== | ||
| Line 265: | Line 264: | ||
* dNTP : 1µL | * dNTP : 1µL | ||
* Phusion : 1µL | * Phusion : 1µL | ||
| - | * | + | * DMS0 : 2µL |
* H2O : 31µL | * H2O : 31µL | ||
Latest revision as of 01:29, 5 October 2013
Notebook : August 14
Lab work
A - Aerobic/Anaerobic regulation system
Objective : characterize BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006
1 - Gel purification of the digestion of BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI
XiaoJing
Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ]
Nanodrop :
- NarG : 10.5ng/µL
- NarK : 16.8ng/µL
- NirB : 24.3ng/µl
|
We lost our plasmids. We will do the digestion again. |
2 - Extraction of BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006 from DH5α
XiaoJing
Protocol : High-copy plamid extraction
Nanodrop :
- NarK: 89.8ng/µL
- NarG : 80.7ng/µL
- Nir B :65.8ng/µL
- Pndh* : 227ng/µL
|
The extraction was good. We will digest BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006. |
3 - Digestion of BBa_K1155004,BBa_K1155005, BBa_K1155006 by EcoRI/SpeI
Nadia
- Buffer FD : 2µL
- H2O : 6µL
- DNA : 10µL
- SpeI FD : 1µL
- EcoRI FD : 1µL
We let the digestion at 37°C during 15 minutes.
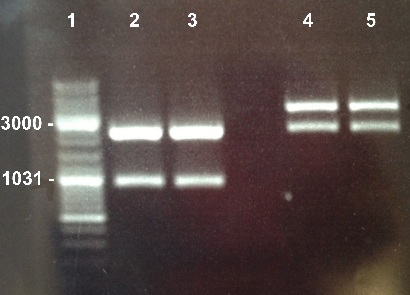
4 - Electrophoresis to check the digestion of BBa_K1155004, BBa_K1155005, BBa_K1155006 by EcoRI/SpeI
Anaïs, Nadia
Expected sizes :
- pSB1C3 : 2070kb
- NarK, NarG, NirB : 200kb
|
We obtained NarK, NarG, NirB fragments at the right size but in very few quantity. We do it again but this time we will use more quantity of enzymes. |
5 - Digestion of BBa_K1155000, BBa_K1155004,BBa_K1155005, BBa_K1155006 by SpeI and EcoRI/SpeI
Anaïs, Nadia
- Digestion by SpeI :
- Buffer : 2µL
- SpeI : 2µL
- ADN : 15µL
- H20 : 1µL
- Digestion by EcoRI and SpeI :
- Buffer : 3µL
- SpeI : 2µL
- EcoRI : 2µL
- ADN : 20µL
- H20 : 3µL
We let digestions at 37°C during 10 minutes.
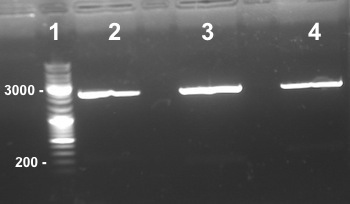
6 - Electrophoresis to check the digestion of BBa_K1155000, BBa_K1155004, BBa_K1155005, BBa_K1155006 by SpeI and EcoRI/SpeI
Anaïs, Nadia
Expected sizes :
- Pndh* in pSB1C3 : 2200bp
- Nar K in pSB1C3, NarG in pSB1C3, NirB in pSB1C3 : 2270bp
- pSB1C3 : 2070kb
- NarK, NarG, NirB : 200kb
- Pndh* : 111bp
|
Well 1 : we have done one digestion so we have to obtain one fragment, we have two stripes, the digestion of BBa_K1155000 by SpeI wasn't good. Well 2, 3, 4, 6, 7 : we obtain NarG, NarK and NirB in pSB1C3 and Pndh*, NarK fragments at the right size, we can purify it. Well 8, 9 : we have done two digestions so we have to obtain two fragments, we have only one stripe, the digestion of BBa_K1155005 and BBa_K1155004 by EcoRI/SpeI wasn't good. |
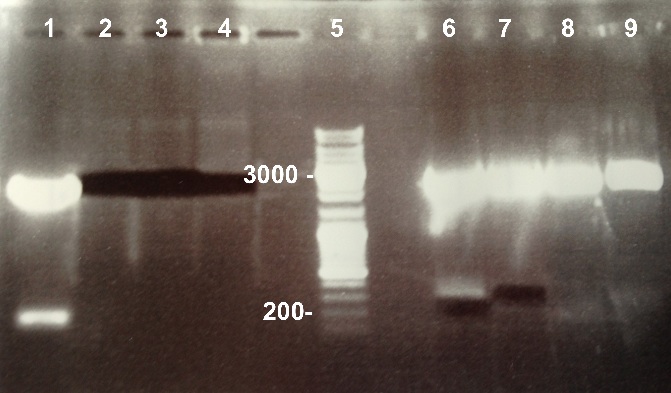
7 - Gel purification of the digestion of BBa_K1155000 and BBa_K1155006 by EcoRI/SpeI and BBa_K1155004, BBa_K1155005 and BBa_K1155006 by SpeI
Anaïs, Nadia
Protocol : [http://www.mn-net.com/tabid/1452/default.aspx Gel purification ]
|
|
Nanodrop :
- Pndh* : 13.7ng/µL
- NarK : 27.9ng/µL
- NarK in PSB1C3 : 87.1ng/µL
- NarG in PSB1C3 : 39.2ng/µL
- NirB in PSB1C3 : 37.7ng/µL
|
The purification was good. We will ligate them. |
8 - Electrophoresis of the digestion of BBa_K1155003, BBa_K1155007 by XBaI/PstI
Nadia
Expected sizes :
- RBS_LacZ-Term : 3500bp
- RBS_AmilCP-Term : 824bp
|
We obtained fragments at the right size. We will purify it. |
9 - Gel purification of the digestion of BBa_K1155003 and BBa_K1155007 by XBaI/PstI
Nadia
Nanodrop :
- RSB-LacZ-Term : 59.6ng/µL
- RBS-AmilCP-Term : 28.7ng/µL
|
We obtained low quantities of plasmid that's why we will make an Ethanol precipitation to concentrate it. |
10 - Glycerol stock of MG1655Z1 Δfnr::Km
XioaJing
Used quantities :
- Glycerol : 500 µL
- MG1655Z1 Δfnr::Km: 1 mL
We stored the bacteria at -20°C.
A - Aerobic/Anaerobic regulation system / B - PCB sensor system
Objective : obtaining FNR and BphR2 proteins
1 - Gibson assembly
Used quantities :
- RBS-BphR2 :
- pSB1C3 : 3µL
- BphR2 Part I : 1µL
- BphR2 Part II : 1µL
- Gibson mix : 15µL
- FNR :
- pSB1C3 : 3µL
- FNR Part I : 1µL
- FNR Part II : 1µL
- Gisbon mix : 15µL
- RBS-FNR :
- pSB1C3 : 3µL
- RBS-FNR Part I : 1µL
- FNR Part II : 1µL
- Gibson mix : 15µL
We incubate these mix at 50°C during 1h inside PCR machine. Then we keep these mix at 4°C during the week end.
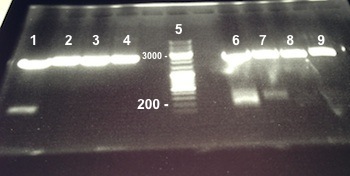
2 - Electrophoresis of PCR products : BphR2 Part I
Damir
|
|
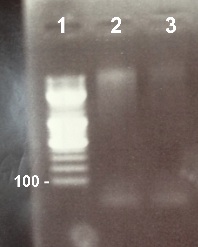
|
|
Expected sizes :
- BphR2 Part I : 178kb
|
On the first gel, all deposits disappear so we did the electrophoresis again. On the second gel, we didn't obtain stripes at the good size. We do the PCR again using new quantities and a new PCR program. |
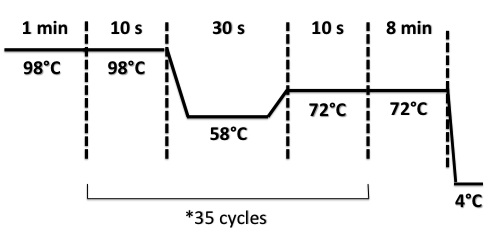
3 - PCR of BphR2 Part I
Damir
Used quantities :
- Oligo 54F : 2µL
- Oligo 55R : 2µL
- DNA : 1µL
- Buffer Phusion : 10µL
- dNTP : 1µL
- Phusion : 1µL
- DMS0 : 2µL
- H2O : 31µL
PCR program :
| Previous day | Back to calendar | Next day |
 "
"