Team:Edinburgh/Introduction/Bioethanol
From 2013.igem.org
Krothschild (Talk | contribs) |
|||
| (11 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div class='content'> | <div class='content'> | ||
| - | + | <h3>Production of bioethanol</h3> | |
| - | Microbial production of ethanol is of great importance due to its possible application as a biofuel. Increasing ethanol yields in bacteria | + | Microbial production of ethanol is of great importance due to its possible application as a biofuel. Increasing ethanol yields in bacteria are potentially beneficial as they are able to utilise a wider variety of renewable, biomass-derived carbon sources compared to the standard ethanol producer: ''Sacharomyces cerevisiae''. |
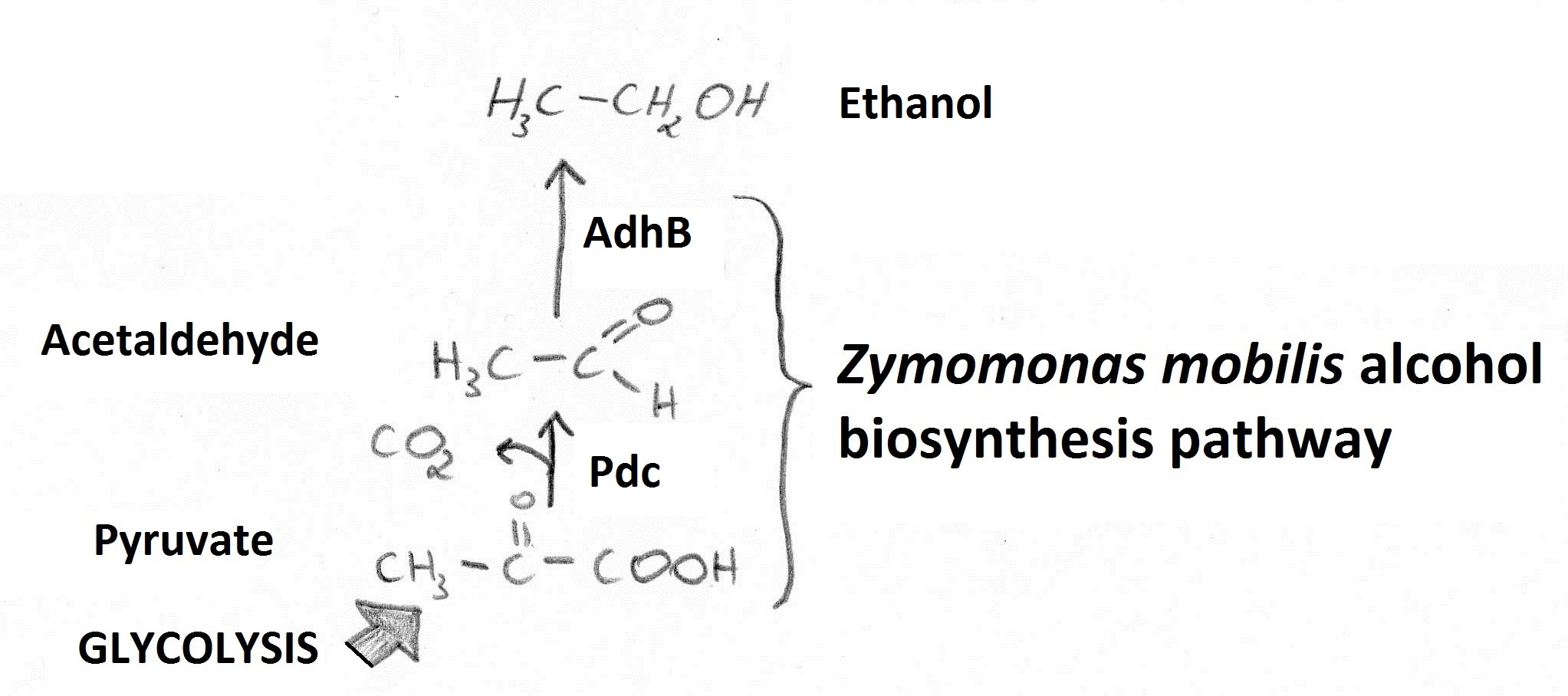
| - | Our goal was to increase ethanol yields using a fusion of Zymomonas mobilis pyruvate decarboxylase (Pdc) and alcohol dehydrogenase B (AdhB). Reactions catalysed by those enzymes (Figure 1) enable conversion of pyruvate to ethanol. | + | Our goal was to increase ethanol yields using a fusion of ''Zymomonas mobilis'' pyruvate decarboxylase (Pdc) and alcohol dehydrogenase B (AdhB). Reactions catalysed by those enzymes (Figure 1) enable conversion of pyruvate to ethanol. |
[[File:bioethanol_introduction_1.jpg|700px]] | [[File:bioethanol_introduction_1.jpg|700px]] | ||
| - | Figure 1. Ethanol is generated from pyruvate (fed into the pathway from glycolysis) via an Acetaldehyde intermediate. | + | '''Figure 1.''' Ethanol is generated from pyruvate (fed into the pathway from glycolysis) via an Acetaldehyde intermediate. |
| - | We based the work on the hypotheses that flow of substrate from one enzyme to another should be facilitated in the fusion protein, and that due to this less of the toxic aldehyde intermediate should be released into the cell. Other fusion proteins were already presented to increase amounts of final product produced (Wang et al., 2011) what further encouraged us to undertake this work. | + | We based the work on the hypotheses that '''flow of substrate from one enzyme to another should be facilitated in the fusion protein''', and that due to this '''less of the toxic aldehyde intermediate should be released''' into the cell. Other fusion proteins were already presented to increase amounts of final product produced (Wang et al., 2011) what further encouraged us to undertake this work. |
We have decided that we will perform the fusion of C terminus of Pdc to N terminus of AdhB. What could potentially generate a protein of a following structure: (figure 2) | We have decided that we will perform the fusion of C terminus of Pdc to N terminus of AdhB. What could potentially generate a protein of a following structure: (figure 2) | ||
| - | [[File:bioethanol_introduction_2. | + | [[File:bioethanol_introduction_2.png|600px]] |
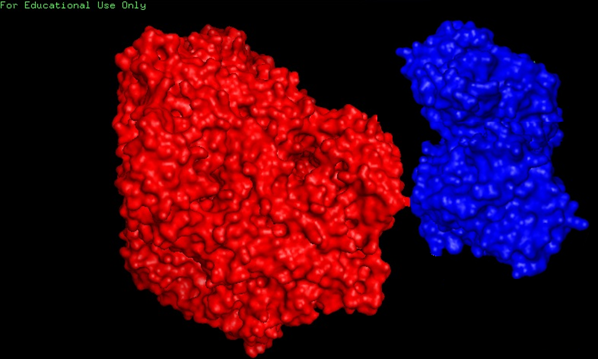
| - | Figure 2. Putative structure of a fused protein. Pdc tetramer (red) is an active state of this enzyme. C terminus of one of the members was linked to an N terminus of AdhB (blue). Presented AdhB enzyme is a dimer and just one of the monomers is linked to Pdc. | + | '''Figure 2.''' Putative structure of a fused protein. Pdc tetramer (red) is an active state of this enzyme. C terminus of one of the members was linked to an N terminus of AdhB (blue). Presented AdhB enzyme is a dimer and just one of the monomers is linked to Pdc. |
| - | We have decided that the most accessible of the waste molecules present in the effluents from the industry would be lactic acid. This molecule is produced as a by- product of anaerobic fermentation and is used in pre-treatment processes within leather | + | We have decided that the most accessible of the waste molecules present in the effluents from the industry would be lactic acid. This molecule is produced as a by- product of anaerobic fermentation and is used in pre-treatment processes within leather and textile industries. Lactic acid can be easily and cheaply purified using various chemical methods (Vijayakumar et al., 2008). |
Using D-lactate dehydrogenase (D-LDH) and L-lactate dehydrogenase (L-LDH) we will be able to generate a metabolic circuit (figure 3) which should directly convert purified lactate into ethanol with high yields. | Using D-lactate dehydrogenase (D-LDH) and L-lactate dehydrogenase (L-LDH) we will be able to generate a metabolic circuit (figure 3) which should directly convert purified lactate into ethanol with high yields. | ||
| Line 27: | Line 27: | ||
[[File:Bioethanol_results10.png]] | [[File:Bioethanol_results10.png]] | ||
| - | + | '''Figure 3.''' Direct conversion of lactic acid to ethanol using LDH and our fusion protein. Note that NADH cofactor is recycled over the course of reaction what directionates the reversible steps catalysed by LDH and ADHB. | |
| - | + | <h2>References:</h2> | |
| - | + | Vijayakumari, J., Aravindan, R. & Viruthagiri, T. 2008. Recent trends in the production, purification and application of lactic acid. Chemical and Biochemical Engineering Quarterly, 22, 245-264. | |
| + | |||
| + | Wang, C., Yoon, S.-H., Jang, H.-J., Chung, Y.-R., Kim, J.-Y., Choi, E.-S. & Kim, S.-W. 2011. Metabolic engineering of Escherichia coli for alpha-farnesene production. Metabolic Engineering, 13, 648-655. | ||
Latest revision as of 01:30, 5 October 2013
Production of bioethanol
Microbial production of ethanol is of great importance due to its possible application as a biofuel. Increasing ethanol yields in bacteria are potentially beneficial as they are able to utilise a wider variety of renewable, biomass-derived carbon sources compared to the standard ethanol producer: Sacharomyces cerevisiae. Our goal was to increase ethanol yields using a fusion of Zymomonas mobilis pyruvate decarboxylase (Pdc) and alcohol dehydrogenase B (AdhB). Reactions catalysed by those enzymes (Figure 1) enable conversion of pyruvate to ethanol.
Figure 1. Ethanol is generated from pyruvate (fed into the pathway from glycolysis) via an Acetaldehyde intermediate.
We based the work on the hypotheses that flow of substrate from one enzyme to another should be facilitated in the fusion protein, and that due to this less of the toxic aldehyde intermediate should be released into the cell. Other fusion proteins were already presented to increase amounts of final product produced (Wang et al., 2011) what further encouraged us to undertake this work.
We have decided that we will perform the fusion of C terminus of Pdc to N terminus of AdhB. What could potentially generate a protein of a following structure: (figure 2)
Figure 2. Putative structure of a fused protein. Pdc tetramer (red) is an active state of this enzyme. C terminus of one of the members was linked to an N terminus of AdhB (blue). Presented AdhB enzyme is a dimer and just one of the monomers is linked to Pdc.
We have decided that the most accessible of the waste molecules present in the effluents from the industry would be lactic acid. This molecule is produced as a by- product of anaerobic fermentation and is used in pre-treatment processes within leather and textile industries. Lactic acid can be easily and cheaply purified using various chemical methods (Vijayakumar et al., 2008).
Using D-lactate dehydrogenase (D-LDH) and L-lactate dehydrogenase (L-LDH) we will be able to generate a metabolic circuit (figure 3) which should directly convert purified lactate into ethanol with high yields.
Figure 3. Direct conversion of lactic acid to ethanol using LDH and our fusion protein. Note that NADH cofactor is recycled over the course of reaction what directionates the reversible steps catalysed by LDH and ADHB.
References:
Vijayakumari, J., Aravindan, R. & Viruthagiri, T. 2008. Recent trends in the production, purification and application of lactic acid. Chemical and Biochemical Engineering Quarterly, 22, 245-264.
Wang, C., Yoon, S.-H., Jang, H.-J., Chung, Y.-R., Kim, J.-Y., Choi, E.-S. & Kim, S.-W. 2011. Metabolic engineering of Escherichia coli for alpha-farnesene production. Metabolic Engineering, 13, 648-655.

| 
| | | | 
|
| This iGEM team has been funded by the MSD Scottish Life Sciences Fund. The opinions expressed by this iGEM team are those of the team members and do not necessarily represent those of MSD | |||||
 "
"