Team:Warsaw/Genetic lab journal
From 2013.igem.org
Tosterovic (Talk | contribs) (→Genetics lab on Pawińskiego street) |
Annamiscicka (Talk | contribs) (→26.07.2013) |
||
| (2 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
''This is jorunal of our genetic lab. [[Team:Warsaw/Cellular biology lab journal|Click here for our cellular biology lab journal.]]'' | ''This is jorunal of our genetic lab. [[Team:Warsaw/Cellular biology lab journal|Click here for our cellular biology lab journal.]]'' | ||
| - | == | + | == I genetic lab (University of Warsaw, Faculty of Biology) == |
====1st week: 2.07.13 – 5.07.13==== | ====1st week: 2.07.13 – 5.07.13==== | ||
| Line 52: | Line 52: | ||
| - | == | + | == II genetic lab (University of Warsaw, Institute of Genetics and Biotechnology) == |
====08.07 – 15.07==== | ====08.07 – 15.07==== | ||
| Line 94: | Line 94: | ||
We extracted plasmids out of E0040, J23100. | We extracted plasmids out of E0040, J23100. | ||
| - | |||
====29. 07.2013==== | ====29. 07.2013==== | ||
| Line 178: | Line 177: | ||
====20.09==== | ====20.09==== | ||
We performed gelout with mOrange PCR product. We isolated DNA from liquid cultures which we created previous day. We also transformed bacteria with pSB1C3. | We performed gelout with mOrange PCR product. We isolated DNA from liquid cultures which we created previous day. We also transformed bacteria with pSB1C3. | ||
| - | Also control digest of isolated DNA was performed. It proved we have failed. We conducted succesful control | + | Also control digest of isolated DNA was performed. It proved we have failed. We conducted succesful control digest of N-terminal and C-terminal ends of GFP. We performed PCR of m6 and m12. |
We digested pSB1C3 and our constructs from GeneRay with EcoRI and PstI. | We digested pSB1C3 and our constructs from GeneRay with EcoRI and PstI. | ||
Latest revision as of 17:28, 4 October 2013
Genetic lab journal
This is jorunal of our genetic lab. Click here for our cellular biology lab journal.
I genetic lab (University of Warsaw, Faculty of Biology)
1st week: 2.07.13 – 5.07.13
Preparation of competent cells for chemical-based transfection. Failure.
2nd week: 8.07.13 – 12.07.13
Successfull preparation of competent cells for chemical-based transfection. Verification with iGEM’s transformation kit. Preparation of competent cells for electroporation. Failure.
3rd week: 15.07.13 – 19.07.13
Chemical-based transformation. BioBricks: J06504, J23100, B0034, I712074, E0040, B0015, B0017, E2020, E2030. Failure: E2020. Succeed: J06504, J23100, B0034, I712074, E0040, B0015, B0017, E2030. Isolate plasmid DNA by alkaline lysis. Quality checked with spectrophotomeric analysis with NanoDrop.
4th week: 22.07.13 – 26.07.13
Transformation with isolated pladmids and BioBricks: I74609 and K864100. Isolate plasmid DNA (from I73609 and K864100) with mini-prep kit. Digest and ligation of construct: J23100 and B0034 on plasmid pSB1C3.
5th week: 29.07.13 – 1.08.13
Preparing construct: J23100 + B0034 + K864100 on plasmid pSB1A3. Measure glowing fluorescent proteins: sfGTP, sfYFP, sfCFP, sfBFP and SYFP2. Superfolded (sf) forms are made by Warsaw Team. SYFP2 is from parts registry.
6th week: 5.08.13 – 9.08.13=
Measuring the fluorescence of obtained proteins. Banking strains with our fluorescent proteins.
7th week: 12.08.13 – 14.08.13
Transformation BioBrick: E2050. PCR: sfBFP N-terminal, sfYFP N-terminal and mCherry (J06504) C-terminal (succeed). Isolate plasmid DNA with: sfGFP (I73609), mOrange (E2050), sfCFP, sfYFP and sfBFP.
8th week: 19.08.13 – 23.08.13
PCR: sfGFP N-terminal (failure), sfCFP N-terminal (succeed) and mCherry N-terminal (succeed). Gel-out: sfYFP N-terminal, sfBFP N-terminal and mCherry C-terminal. Cloning sfCFP N-terminal, mCherry N-terminal, sfYFP N-terminal, sfBFP N-terminal and mCherry C-terminal to pSB1C3 plasmid and inserting it to bacteria. Preparing construct: J23100+B0034+E0040 – failure.
9th week: 26.08.13 – 30.08.13
Verification cloning
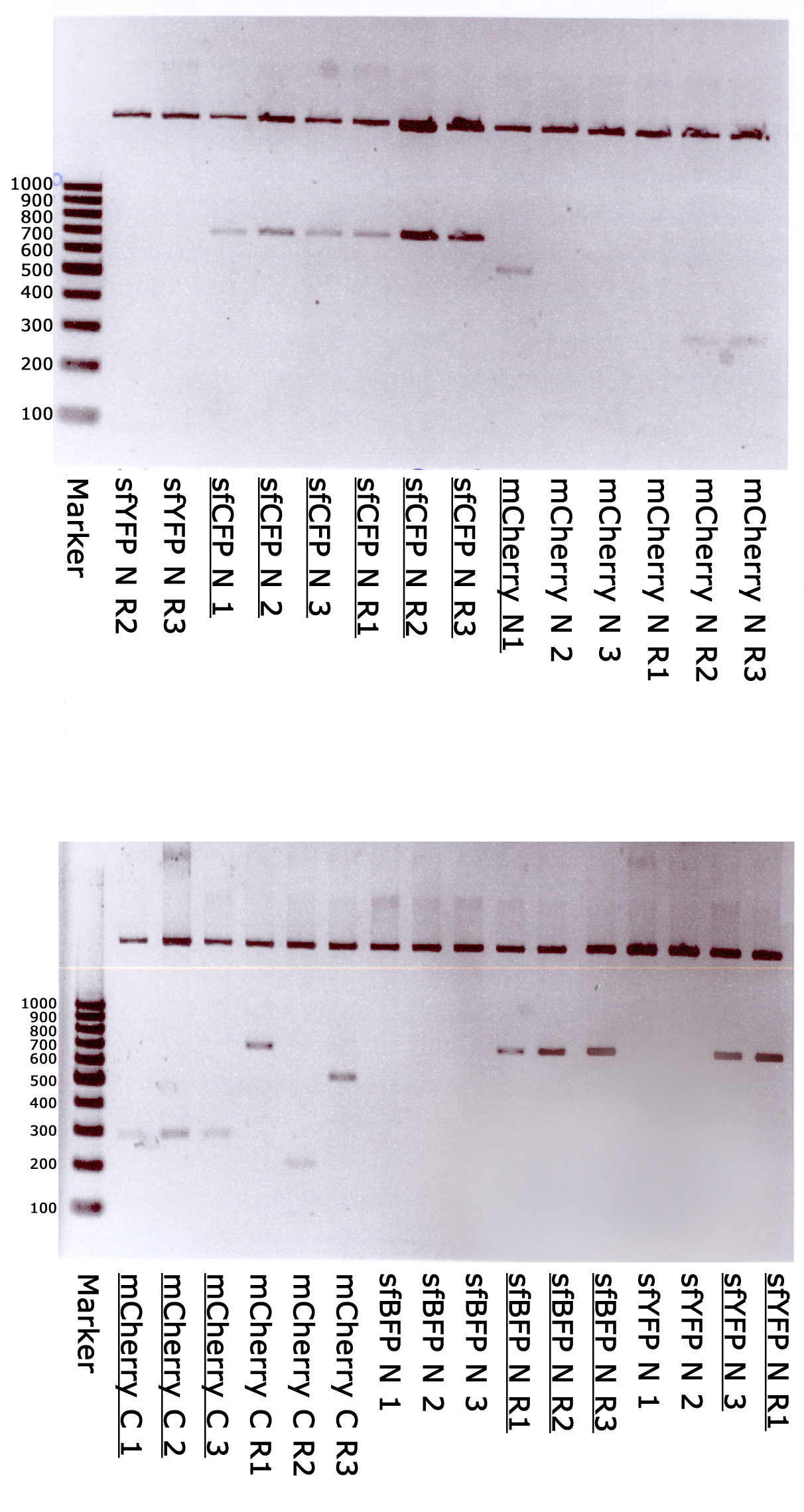
Marker - GeneRuler 100 bp DNA Ladder, ready-to-use (Thermo); sfYFP N 1,2,3,R1,R2,R3 - random clones of sfYFP N-terminal; sfCFP N 1,2,3,R1,R2,R3 - random clones of sfCFP N-terminal; mCherry N 1,2,3,R1,R2,R3 - random clones of mCherry N-terminal; mCherry C 1,2,3,R1,R2,R3 - random clones of mCherry C-terminal; sfBFP N 1,2,3,R1,R2,R3 - random clones of sfBFP N-terminal. Underlined - correct clones.
And great success! PCR: sfGFP N-terminal and GFP C-terminal (various attenuation template DNA) and sfC-terminal (to versions: m6 and m12 to all of superfolded fluorescent proteins). Cloning and inserting it to bacteria. Once again preparing construct: J23100+B0034+E0040.
10th week: 2.09.13 – 7.09.13
Welcome school! Verification cloning. Unfortunately epic fail. PCR: sfGFP N-terminal, GFP N-terminal and GFP C-terminal. At least success!
11th week: 10.09.13 – 14.09.13
Cloning sfGFP N-terminal, GFP N-terminal and GFP C-terminal. Unfortunately fail. Checking buffers for digesting – they’re fine. Searching mistakes in cloning.
12th week: 16.09.13 – 20.09.13
We get out commission from GeneRay. Cloning this to pSB1C3 plasmids, but it ends with failure. Cleaning lab and end work.
II genetic lab (University of Warsaw, Institute of Genetics and Biotechnology)
08.07 – 15.07
We isolated plasmids from the following probes: G, Y1, Y2, Y3, B1, B2, B3. Then we conducted a control digest. Only probes Y1, Y2 and Y3 contained plasmids we wanted and we sent those for sequencing. It showed that probe Y2 contained correct plasmid encoding sfYFC protein. We made long term stock from Y1, Y2 and Y3 probes. Afterwards we performed the whole procedure once again to isolate plasmids we were not able to obtain previously. We successfully isolated plasmid that encodes sfCFP and we sent it for sequencing. Sequencing ensured us that we isolated the correct one. We also made electrocompetent bacteria for our further experiments. We conducted dialysis of our ligated sfGFP, sfCFP, sfYFP and sfBFP (variants with terminator) and used these plasmids to transform electrocompetent bacteria. We were able to successfully transform them with plasmids that encode sfBFP and sfYFP. We isolated these plasmids and sent them for sequencing.
17.07.2013
We wanted to obtain BFP on pSB1C plasmid for parts registry and CFP on J231CO for testing purposes. To minimize possibility of failure we did it in two different ways: by directed mutagenesis and by ligation of proper insert with plasmid.
We performed mutagenesis using PCR. We started two PCR reactions: one with CFP on pSB1C as a template and primers introducing mutation to BFP, second with BFP on J231CO as template and primers introducing mutation to CFP. We put 20 microliters of each mix and started reaction with 58C annealing temperature and 4 minutes for complementary strand synthesis.
18.07.2013
From PCR reaction products: BFP on pSB1C and CFP on J231CO we digested template plasmid using DPN1. Afterwards we performed dialysis and transformed bacteria with mutation product by electroporation. After 60 minutes in 37C bacteria were moved on plates with L1 mixed with chloramphenicol and ampicillin respectively.
We started ligation of psB1C with BFP and J231CO with CFP.
19.07.2013
We transformed bacteria with ligation product from yesterday. Afterwards we put the bacteria on plates with proper antibiotic.
We created culture fluid from BFP on pSB1C from plates obtained from mutation. We put selected strains from plate into 5 probes containing 5ml of L1 and 5 microliters of chloramphenicol. We performed streaking of CFP on J231CO on 6 plates with ampicillin.
22.07.2013
Bacteria transformed with ligation product didn’t grow on plates from 19.07.
We created the culture fluid out of the two CFP on J231CO plates. We extracted plasmids from 5 probes with BFP on pSB1C and measured extracts with nanodrop. We also transformed bacteria with 9 plasmids: and put them on plates with chloramphenicol.
23. 07. 2013
We isolated plasmids from bacteria containing sfCFP genes (probes C2 and C4). Plasmids were sent for sequencing. We also created plate cultures with E2020, E0040, B0034, I712074 and J23100.
24. 07. 2013
We created fluid cultures from plate cultures that have grown (B0034 and I712074). We also repeated transforming bacteria with plasmids E0040, E2020, J23100 and created plate cultures from them.
25. 07. 2013
We extracted plasmids E2030, B0017, J06504, J712074 and I2310. We also started fluid cultures from plates that contained E0040, E2020, J23100.
26.07.2013
We started digest of B2 on pSB1C with PstI and EcorI in Red buffer to obtain the plasmid and digest of YFP, BFP and CFP to get the inserts. Aim is to obtain plasmids with chloramphenicol resistance that can be expressed. After a day in 37C E0040 and J23100 fluid cultures grew while E2020 did not. There were newly grown colonies on E2020 plate so we started a new fluid culture from one of them.
We extracted plasmids out of E0040, J23100.
29. 07.2013
We tried to perform gel electrophoresis with digest products but it was unsuccessful - probably digest failed as it was left over for 2 nights. We again performed digest of B5 with EcoR1 and Pst1 to obtain psB1C in Orange buffer and YFP, BFP and CFP. We transformed bacteria with YFP, BFP and CFP on J231CO plasmid and put them on the plates.
02.08.2013
We performed PCR of BIFC N-end YFP, BFP, CFP.
05.08.2013
We put the bacteria transformed with YFP, BFP and CFP on psB1C plasmids (expressive parts) on plates with chloramphenicol. We started digest of B4 to obtain psB1C in Orange buffer with EcoR1 and Pst1. We performed electrophoresis of PCR product from 02.08 and then we extracted the product from the gel. We started the digest of extracts with XbaI and BcuI (SpeI - isoschizomer which cannot be deactivated by heat, therefore we have to put it through DNA purification procedure).
We performed PCR of BIFC N-end GFP and put it to the digest with XbaI, BcuI
06.08.2013
We purified the digested PCR product from 02.08 but with poor results. We started fluid culture of YFP, BFP and CFP on chloramphenicol and performed next PCR of Bifc N-parts: YFP, BFP, CFP and GFP.
07.08.2013
We extracted plasmids from fluid culture. We put bacteria from these fluid cultures on plates to check if they are correct. We performed electrophoresis of psB1C3 digested plasmid and PCR product from yesterday.
13.08.2013
We extracted pSB1C3 plasmid and performed its ligation with mCherry 1 and mCherry 2.
14.08.2013
We performed dialysis of ligated psB1C3 with mCherry (both of them). We transformed bacteria with these plasmids and then we put them on plates. We also extracted CFP on J23160 using gel electrophoresis. We used it to transform bacteria and then we put them on plates.
15.08.2013
We started fluid culture of bacteria with CFP and mCherry.
16.08.2013
We isolated CFP from bacteria and performed a digest. We also isolated mCherry, performed a control digest, which unfortunately proved we failed.
19.08.2013
We extracted pSB1C3 plasmid using gel electrophoresis (gelout). We ligated previously digested CFP with obtained pSB1C3.
20.08.2013
Dialysis of ligated CFP was performed. Afterwards we transformed bacteria with it and then we put them on plates. That day we also started 2 fluid cultures. One in order to get electrocompetent bacteria. Second to obtain pSB1C3 plasmid.
21.08.2013
Isolation of pSB1C3 and its digest.
22.08.2013
We started fluid culture of CFP and grew electrocompetent bacteria.
23.08.2013
We isolated CFP, performed its control digest and gel electrophoresis. We put electrocompetent bacteria on plates to measure their efficiency.
03.09.2013
We made some new plates. We also measured efficiency of electrocompetent bacteria. We tried performing mutagenesis of CFP into BFP using PCR.
04.09.2013
We performed purification of PCR product, transformed bacteria with it and then put them on plates. Hopefully they contain BFP. We performed PCR of genes encoding C-terminal and N-terminal ends of GFP and PCR of genes encoding C-terminal end of the rest of our superfolder fluorescent proteins.
10.09.2013
We performed PCR of genes with C-terminal and N-terminal ends of GFP because we lost the previous PCR products due to mistake. We performed control gel electrophoresis of obtained PCR product. The results were correct. We started fluid culture of bacteria with BFP. We isolated pSB1C3 from the stock bacteria.
11.09.2013
We stared fluid culture of CFP (to make long term stock of them), YFP (because we lost isolated plasmids) and BFP. Afterwards we tried to isolate plasmids with BFP. First attempt was a failure. Next we performed gel electrophoresis and the results were poor as well. Finally we used alkaline lysis and it was a success. That day we also used gel electrophoresis and a clean-up kit to purify the PCR product containing C-terminal and N-terminal GFP ends. At the end we performed ligation of m6 and m12 with pSB1C3 plasmid.
12.09.2013
We made long term stock of bacteria with CFP. We isolated plasmids with YFP and started another fluid culture of bacteria containing it. We performed dialysis of plasmids with m6 and m12 constructs. Then we digested genes encoding C-terminal and N-terminal GFP ends. We designed primers for mOrange PCR.
13.09.2013
We started fluid culture of bacteria with m6 and m12 genes. We ligated pSB1C3 plasmid with genes DNA encoding C-terminal and N-terminal ends of GFP. We isolated pSB1C3 plasmid.
16.09.2013
We failed to isolate pSB1C3 plasmid and plasmids with m6 and m12. We performed dialysis of plasmids with C-terminal and N-terminal ends of GFP. We transformed bacteria with it and put them on plates. This day we got synthesized parts we ordered. We transformed bacteria with them and put them on plates. We digested obtained parts with EcoRI and PstI.
17.09.2013
We performed isolation of m6 and m12 parts and pSB1C3 plasmid. We started fluid cultures from synthesized parts and performed their ligation with pSB1C3 plasmid.
18.09.2013
We performed dialysis of synthesized parts ligated with plasmids. We transformed bacteria with them and put the bacteria on plates. We started fluid cultures of these parts. We also started fluid cultures of: m6 and m12 (8 of each); HBB6, HBB12 and HBA (3 of each). We isolated plasmids with synthesized parts (as spare copies). We also isolated C-GFP and N-GFP plasmids. We performed control digest of C-GFP and N-GFP with EcoRI and PstI. We performed PCR of mOrange part.
19.09.2013
We started fluid cultures of synthesised parts from GeneRay. We tried isolation of m6 and m12 which failed (we used Fermentas kit). We repeated isolation of m6 and m12 which again was a failure. We also checked using gel electrophoresis that PCR of mOrange was successful.
20.09
We performed gelout with mOrange PCR product. We isolated DNA from liquid cultures which we created previous day. We also transformed bacteria with pSB1C3. Also control digest of isolated DNA was performed. It proved we have failed. We conducted succesful control digest of N-terminal and C-terminal ends of GFP. We performed PCR of m6 and m12. We digested pSB1C3 and our constructs from GeneRay with EcoRI and PstI.
21.09
We performed unssucesful gelout of m6 and m12 (PCR products). We ligated GeneRay constructs and N, C terminal ends of GFP with pSB1C3. We transformed bacteria with all these constructs and created a plate cultures with them. We also performed PCR of m6 and m12
22.09
We digested pSB1C3 with EcoRI and PstI. We performed clean up on m6 and m12 PCR products.
23.09
Control digest of m12 and m6 PCR products confirmed failure of PCR.
24.09
We performed PCR of GeneRay constructs.
27.09
We performed control electrophoresis of GeneRay constructs PCR products. It confirmed failure. We also performed gradient PCR of m6 and m12
28.09
We conducted control electrophoresis of m6 and m12 PCR products. It ensured us we failed.
 "
"