Team:Paris Saclay/Notebook/July/11
From 2013.igem.org
| (One intermediate revision not shown) | |||
| Line 5: | Line 5: | ||
=='''Lab work'''== | =='''Lab work'''== | ||
| - | ===='''Objective : obtaining obtaining biobricks in | + | ===='''Objective : obtaining obtaining biobricks in pSB3K3'''==== |
===='''1 - Extraction of BBa_J04450 from DH5α'''==== | ===='''1 - Extraction of BBa_J04450 from DH5α'''==== | ||
| Line 87: | Line 87: | ||
===='''Objective : obtaining BBa_K1155001, BBa_K1155002, BphR2'''==== | ===='''Objective : obtaining BBa_K1155001, BBa_K1155002, BphR2'''==== | ||
| + | |||
===='''1 - Colony PCR of BBa_K1155001, BBa_K1155002 and BphR2 in pSB1C3 in DH5α to check good insertions of BphR1, BphA1, BphR2 in pSB1C3'''==== | ===='''1 - Colony PCR of BBa_K1155001, BBa_K1155002 and BphR2 in pSB1C3 in DH5α to check good insertions of BphR1, BphA1, BphR2 in pSB1C3'''==== | ||
| Line 191: | Line 192: | ||
Expected sizes : | Expected sizes : | ||
| - | *BphR2_Up/VR, VF/BphR2_Down : 1200bp | + | * BphR2_Up/VR, VF/BphR2_Down : 1200bp |
{| | {| | ||
| Line 204: | Line 205: | ||
Expected sizes : | Expected sizes : | ||
| - | *BphA1_Up/VR, VF/BphA1_Down : 500bp | + | * BphA1_Up/VR, VF/BphA1_Down : 500bp |
{| | {| | ||
| Line 210: | Line 211: | ||
| style="width:350px;border:1px solid black;vertical-align:top;" | | | style="width:350px;border:1px solid black;vertical-align:top;" | | ||
* Well 1 : 6µL of DNA Ladder | * Well 1 : 6µL of DNA Ladder | ||
| - | * Well 2 to | + | * Well 2 to 5 : 5µL of BphR1 with VF/VR primers+1µL of 6X loading dye |
| - | * Well | + | * Well 6 to 13 : 5µL of BphR2 with VF/VR primers+1µL of 6X loading dye |
| - | * Well | + | * Well 14 : 6µL of DNA Ladder |
| + | * Well 15 to 22 : 5µL of BphA1 with VF/VR primers+1µL of 6X loading dye | ||
* Well 17 : 6µL of DNA Ladder | * Well 17 : 6µL of DNA Ladder | ||
* Gel : 1.5% | * Gel : 1.5% | ||
Latest revision as of 23:58, 4 October 2013
Notebook : July 11
Lab work
Objective : obtaining obtaining biobricks in pSB3K3
1 - Extraction of BBa_J04450 from DH5α
Sheng
|
Mini and maxi preparation of 07/10/13 works. We will extract DNA. |
Protocol : Low copy plamid extraction
2 - Digestion of BBa_J04450 to chek the size for the plasmid
Sheng
Used quantities :
- XhoI :
- DNA : 3µL
- Buffer Green : 3µL
- XhoI : 1µL
- H2O : 21µL
- SacII :
- DNA : 3µL
- Buffer Blue : 3µL
- SacII : 1µL
- H2O : 21µL
- EcoRI/PstI :
- DNA : 5µL
- Buffer Orange : 3µL
- EcoRI : 1µL
- PstI : 1µL
- H2O : 20µL
- XhoI/SacII :
- DNA : 5µL
- Buffer Green : 3µL
- XhoI : 1µL
- SacII : 1µL
- H2O : 20µL
3 - Electrophoresis of the digestion of BBa_J04450
Zhou
Expected size
- pSB3K3 : 3819 bp
- EcoRI/PstI : 1069 bp + 2750 bp
- XhoI : 2976 bp + 843 bp
- SacII : 3819 bp
- XhoI/SacII : 843 bp + 616 bp + 2367 bp
|
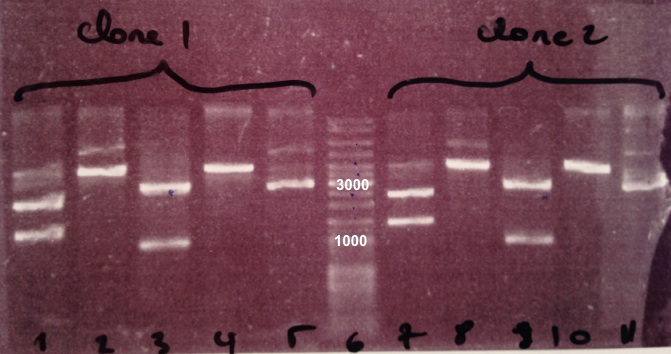
We obtain fragments at the right size for pSB3K3, EcoRI/PstI digestion and SacII digestion. Digestoins with XhoI didn't seem to work. The extraction of BBa_J04450 in DH5α was good. |
B - PBC sensor system
Objective : obtaining BBa_K1155001, BBa_K1155002, BphR2
1 - Colony PCR of BBa_K1155001, BBa_K1155002 and BphR2 in pSB1C3 in DH5α to check good insertions of BphR1, BphA1, BphR2 in pSB1C3
Abdou, Anaïs, Zhou
|
Transformation of BBa_K1155001, BBa_K1155002 and BphR2 protein in DH5α of 07/10/13 work. We will do a Colony PCR. |
We mix our colonies in 10µL of H2O.
Used quantities :
- DNA : 2µL
- Mix A : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- BphR1_Up/VR : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix B : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- VF/BphR1_Down : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix C : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- BphR2_Up/VR : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix D : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- VF/BphR2_Down : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix E : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- BphA1_Up/VR : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix F : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 50µL
- MgCl2 : 20µL
- dNTP : 10µL
- VF/BphA1_Down : 1.25µL for each oligo
- Enzyme : 2.5µL
- H2O : 145µL
- Mix G : (it was divided in 8tubes for 8 colonies different)
- Buffer Go Taq : 125µL
- MgCl2 : 50µL
- dNTP : 25µL
- VF/VR: 3µL for each oligo
- Enzyme : 6.25µL
- H2O : 362.75µL
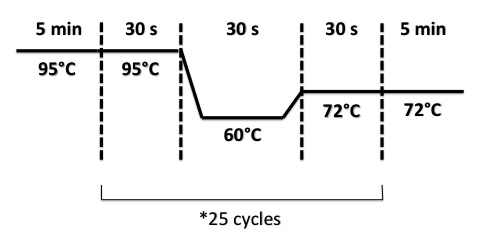
PCR program :
2 - Electrophoresis of Colony PCR products : BBa_K1155001, BBa_K1155002 and BphR2 in pSB1C3
Abdou, Anaïs
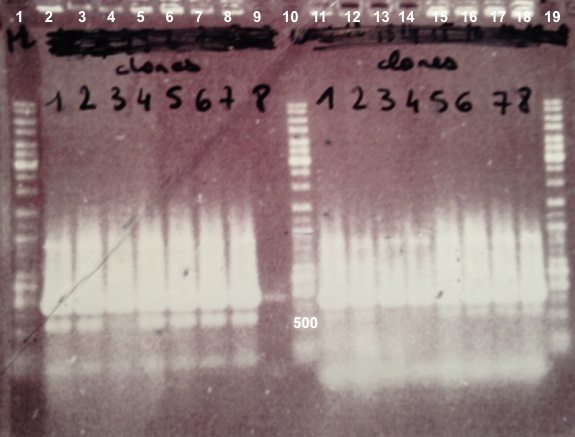
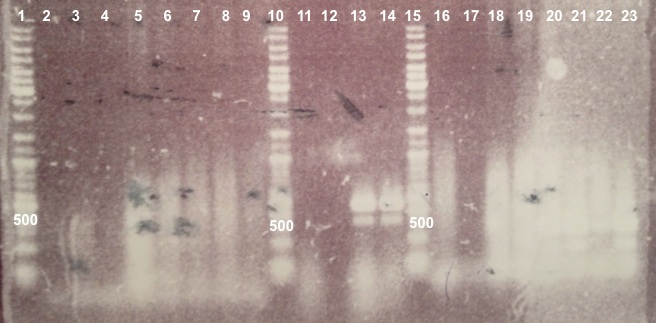
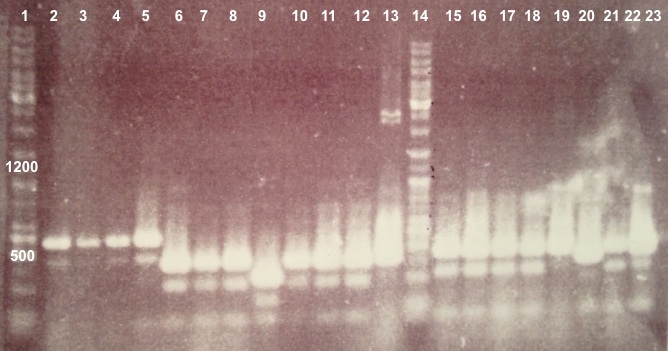
Expected sizes :
- BphR1_Up/VR, VF/BphR1_Down : 500bp
Expected sizes :
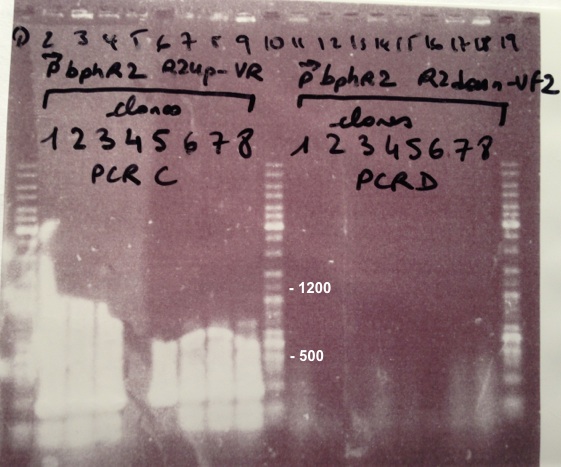
- BphR2_Up/VR, VF/BphR2_Down : 1200bp
Expected sizes :
- BphA1_Up/VR, VF/BphA1_Down : 500bp
Expected sizes :
- BphR2 : 1200bp
- BphR1, BphA1 : 500bp
|
We didn't obtain fragments at the right size but we will do streaking of clone 5, 6 for BphR1, clone 5, 6, 7, 8 for BphA1, clones 3, 4 for BphR2. |
| Previous day | Back to calendar | Next day |
 "
"