|
|
| (70 intermediate revisions not shown) |
| Line 6: |
Line 6: |
| | <!-- Start of content --> | | <!-- Start of content --> |
| | | | |
| - | == Creation of transgenic ''Physcomitrella patens'' plants== | + | ==The PhyscoFilter for Erythromycin== |
| - | | + | |
| - | [[File:TUM13_Workflow_Transformation.png|thumb|center|910px|'''Figure 1:''' Work flow for the generation of transgenic moss.]]
| + | |
| - | | + | |
| - | ==1. Generation of expression constructs==
| + | |
| - | | + | |
| - | {|cellspacing="0" border="1" right
| + | |
| - | |+ '''Table 1:''' Transgenic ''Physcomitrella patens'' plants
| + | |
| - | !Number
| + | |
| - | !Construct name (abbreviation)
| + | |
| - | !BioBrick
| + | |
| - | !GM-Moss
| + | |
| - | |-
| + | |
| - | |PF-1
| + | |
| - | |GFP (GFP-cyt)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159311 BBa_K1159311]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-2
| + | |
| - | |Igk-GFP (GFP-sec)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159304 BBa_K1159304] + [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159311 BBa_K1159311]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-3
| + | |
| - | |NanoLuciferase (nLuc-cyt)
| + | |
| - | |<partinfo>BBa_K1159001</partinfo>
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-4
| + | |
| - | |Igk-NanoLuciferase (nLuc-sec)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159006 BBa_K1159006]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-5
| + | |
| - | |SERK-NanoLuciferase (SERK-nLuc)
| + | |
| - | |<partinfo>BBa_K1159010</partinfo>
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-6
| + | |
| - | |NanoLuciferase-Receptor (nLuc-rec)
| + | |
| - | |<partinfo>BBa_K1159015</partinfo>
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-7
| + | |
| - | |Erythromycinesterase (EreB-cyt)
| + | |
| - | |<partinfo>BBa_K1159000</partinfo>
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-8
| + | |
| - | |Erythromycinesterase-Receptor (EreB-rec)
| + | |
| - | |<partinfo>BBa_K1159014</partinfo>
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-9
| + | |
| - | |Ig Kappa Erythromycinesterase (EreB-sec)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159005 BBa_K1159005]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-10
| + | |
| - | |Laccase-Receptor (Lac-rec)
| + | |
| - | |<partinfo>BBa_K1159016</partinfo>
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-11
| + | |
| - | |Ig Kappa Laccase (Lac-sec)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159007 BBa_K1159007]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-12
| + | |
| - | |Catecholdioxygenase (XylE-cyt)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159012 BBa_K1159012]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-13
| + | |
| - | |DDT-dehydrochlorinase (GST-cyt)
| + | |
| - | |<partinfo>BBa_E0040</partinfo>
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-14
| + | |
| - | |PP1-Receptor (PP1-rec)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159019 BBa_K1159019]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-15
| + | |
| - | |FluA-Receptor (FluA-rec)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159017 BBa_K1159017]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-16
| + | |
| - | |Stressinducible_Promoter-RFP (Stress)
| + | |
| - | |<partinfo>BBa_E0040</partinfo>
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-17
| + | |
| - | |SpyCatcher-Receptor:Spytag-nLuc (Catcher:Tag-nLuc)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159212 BBa_K1159212]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-18
| + | |
| - | |SpyCatcher-Receptor:nLuc-Spytag (Catcher:nLuc-Tag)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159213 BBa_K1159213]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-19
| + | |
| - | |SpyTag-Receptor:SpyCatcher-nLuc (Tag:Catcher-nLuc)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159214 BBa_K1159214]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-20
| + | |
| - | |SpyTag-Receptor:nLuc-SpyCatcher (Tag:nLuc-Catcher)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159215 BBa_K1159215]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-21
| + | |
| - | |Alcohol acetyltransferase I (Banana)
| + | |
| - | |[http://parts.igem.org/Part:BBa_J45014 BBa_J45014]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-22
| + | |
| - | |Kill-switch with PIF3 (PIF-3)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159107 BBa_K1159107]
| + | |
| - | |no
| + | |
| - | |-
| + | |
| - | |PF-23
| + | |
| - | |Kill-switch with PIF6 (PIF-6)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159108 BBa_K1159108]
| + | |
| - | | no
| + | |
| - | |-
| + | |
| - | |PF-24
| + | |
| - | |Kill-switch with PIF6 (FRET)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159120 BBa_K1159120]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |PF-25
| + | |
| - | |Kill-switch with PIF3 (FRET)
| + | |
| - | |[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159119 BBa_K1159119]
| + | |
| - | |<html><img src="https://static.igem.org/mediawiki/2013/0/09/TUM13_checkmark.png" width="25px" class="checkmark"></img></html>
| + | |
| - | |-
| + | |
| - | |}
| + | |
| - | | + | |
| - | ==2. Preparation of linear DNA==
| + | |
| - | In order to transfrom the moss, the DNA including our BioBricks with the nptII selection casette had to be linearized. We used EcoRI for the preparative digestion of our midipreps and simultaneously ran an analytical digest as quality control. Our preparations continued by purifying our digested DNA through salt, isopropanol and ethanol precipitation, solubilising in a small amount of water, measuring the concentration at the NanoDrop and suspending it in different volumes of sterile filtered Ca(NO<sub>3</sub>)<sub>2</sub> buffer for the intended concentration of 0.25 µg/µl .
| + | |
| - | [[File:TUM13_Midiprep_picture1.png|thumb|left|450px|'''Figure 2''': Louise and Christopher preparing midipreps]]
| + | |
| - | [[File:TUM13 Midiprep picture2.png|thumb|right|420px|'''Figure 3''': Midipreps for the moss transformation]]
| + | |
| - | [[File:TUM13_Midiprepgel.png|thumb|center|910px|'''Figure 4''': Analytical gelelectrophoresis of linearized DNA constructs]]
| + | |
| - | | + | |
| - | ==3. Transformation of ''Physcomitrella patens''==
| + | |
| - | To transform Physcomitrella patens, the moss material has to be taken from the liquid culture and its cell walls have to be digested with driselase dissolved in mannitol to obtain protoplasts. The protoplasts are isolated by passing the digested material through sieves and the enzyme is washed off with mannitol and then resuspended in mannitol.
| + | |
| - | | + | |
| - | The number of protoplasts is determined with a hemocytometer and the material is suspended in the right amount of 3M medium to adjust the concentration. The linearized and purified DNA is mixed with PEG4000 and the protoplast solution and incubated while regularly mixing. After incubation, the mixture is diluted with 3M medium, centrifuged and resuspended in regeneration medium.
| + | |
| - | | + | |
| - | The protoplasts are put into 6-well plates, left in the dark over night and then left for 10 days for the regeneration of the cell walls at standard conditions. After moving the protoplasts onto solid medium covered with a layer of cellophane for three days, they are transferred to solid selection medium plates for two weeks. To ensure stabile integration, repeat the two weeks of selection after a two week release phase.
| + | |
| - | | + | |
| - | ===Trips to Freiburg ===
| + | |
| - | <html>
| + | |
| - | <div class="box-center">
| + | |
| - | <ul class="bxgallery">
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/4/48/TUM13_Foto_Freiburgvisit1.jpg/350px-TUM13_Foto_Freiburgvisit1.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/4/41/TUM13_Foto_Freiburgvisit2.jpg/350px-TUM13_Foto_Freiburgvisit2.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/d/d8/TUM13_Foto_Freiburgvisit3.jpg/350px-TUM13_Foto_Freiburgvisit3.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/25/TUM13_Foto_Freiburgvisit4.jpg/350px-TUM13_Foto_Freiburgvisit4.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/1/13/TUM13_Foto_Freiburgvisit5.jpg/350px-TUM13_Foto_Freiburgvisit5.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/5/50/TUM13_Foto_Freiburgvisit6.jpg/350px-TUM13_Foto_Freiburgvisit6.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/27/TUM13_Foto_Freiburgvisit7.jpg/350px-TUM13_Foto_Freiburgvisit7.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/4/46/TUM13_Foto_Freiburgvisit8.jpg/350px-TUM13_Foto_Freiburgvisit8.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/d/df/TUM13_Foto_Freiburgvisit9.jpg/350px-TUM13_Foto_Freiburgvisit9.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/2e/TUM13_Foto_Freiburgvisit10.jpg/350px-TUM13_Foto_Freiburgvisit10.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/26/TUM13_Foto_Freiburgvisit11.jpg/350px-TUM13_Foto_Freiburgvisit11.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/d/de/TUM13_Foto_Freiburgvisit12.jpg/350px-TUM13_Foto_Freiburgvisit12.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/3/3b/TUM13_Foto_Freiburgvisit13.jpg/350px-TUM13_Foto_Freiburgvisit13.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/e/ec/TUM13_Foto_Freiburgvisit14.jpg/350px-TUM13_Foto_Freiburgvisit14.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/e/e6/TUM13_Foto_Freiburgvisit15.jpg/350px-TUM13_Foto_Freiburgvisit15.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/7/70/TUM13_Foto_Freiburgvisit16.jpg/350px-TUM13_Foto_Freiburgvisit16.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/8/8e/TUM13_Foto_Freiburgvisit17.jpg/350px-TUM13_Foto_Freiburgvisit17.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/b/b9/TUM13_Foto_Freiburgvisit18.jpg/350px-TUM13_Foto_Freiburgvisit18.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/9/94/TUM13_Foto_Freiburgvisit19.jpg/350px-TUM13_Foto_Freiburgvisit19.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/f/fc/TUM13_Foto_Freiburgvisit20.jpg/350px-TUM13_Foto_Freiburgvisit20.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/5/54/TUM13_Foto_Freiburgvisit21.jpg/350px-TUM13_Foto_Freiburgvisit21.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/5/5a/TUM13_Foto_Freiburgvisit22.jpg/350px-TUM13_Foto_Freiburgvisit22.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/2/23/TUM13_Foto_Freiburgvisit23.jpg/350px-TUM13_Foto_Freiburgvisit23.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/9/91/TUM13_Foto_Freiburgvisit24.jpg/350px-TUM13_Foto_Freiburgvisit24.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/8/81/TUM13_Foto_Freiburgvisit25.jpg/350px-TUM13_Foto_Freiburgvisit25.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/f/f9/TUM13_Foto_Freiburgvisit26.jpg/350px-TUM13_Foto_Freiburgvisit26.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/3/3b/TUM13_Foto_Freiburgvisit27.jpg/350px-TUM13_Foto_Freiburgvisit27.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/f/f2/TUM13_Foto_Freiburgvisit28.jpg/350px-TUM13_Foto_Freiburgvisit28.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/0/0d/TUM13_Foto_Freiburgvisit29.jpg/350px-TUM13_Foto_Freiburgvisit29.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/a/a7/TUM13_Foto_Freiburgvisit30.jpg/350px-TUM13_Foto_Freiburgvisit30.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/a/ab/TUM13_Foto_Freiburgvisit31.jpg/350px-TUM13_Foto_Freiburgvisit31.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/3/3d/TUM13_Foto_Freiburgvisit32.jpg/350px-TUM13_Foto_Freiburgvisit32.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/0/06/TUM13_Foto_Freiburgvisit33.jpg/350px-TUM13_Foto_Freiburgvisit33.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/1/12/TUM13_Foto_Freiburgvisit34.jpg/350px-TUM13_Foto_Freiburgvisit34.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/3/31/TUM13_Foto_Freiburgvisit35.jpg/350px-TUM13_Foto_Freiburgvisit35.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/1/16/TUM13_Foto_Freiburgvisit36.jpg/350px-TUM13_Foto_Freiburgvisit36.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/9/95/TUM13_Foto_Freiburgvisit37.jpg/350px-TUM13_Foto_Freiburgvisit37.jpg" /></li>
| + | |
| - | <li><img src="https://static.igem.org/mediawiki/2013/thumb/a/ac/TUM13_Foto_Freiburgvisit38.jpg/350px-TUM13_Foto_Freiburgvisit38.jpg" /></li>
| + | |
| - | </ul>
| + | |
| - | </div>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | We had the great chance to perform our '' '''Physcomitrella patens''' '' transformations at Prof. Dr.Reski´s lab in Freiburg, with Dr. Wiedemann as our expert instructor. Our first trip started in a great hurry, because we worked on our DNA preparations until the very last minute. Ingmar even pulled an all-nighter to get the DNA ready. We would have missed our intercity bus if it wasn’t for Rosario who drove us to the bus station, all squished together in the “pizza mobile” with the trunk full with our medium bottles and lab equipment. After five hours on the bus, we fell into bed to get some rest, because we had a very long day ahead of us.
| + | |
| - | | + | |
| - | We arrived at the Reski lab early in the morning to meet with Dr. Gertrud Wiedemann, who instructed us throughout the day and gave us many tips how to proceed with the moss. Because of the incubation times and because it was our first try, it took us ten hours without a break until we had two boxes stacked with 6 well plates. We quickly went to get some beers and chips and met with the Freiburg iGEM team for a really nice barbecue. When we finally left, public transport wasn’t running anymore, so we didn’t miss the chance to take a midnight sightseeing tour through Freiburg, where Volker showed us around.
| + | |
| - | | + | |
| - | Ten days later, Johanna and Andi visited the Freiburg lab to transfer the then regenerated protoplasts onto agar plates and soon after, we came back for our second and final round of transformation. At our first try, we didn’t get enough moss protoplasts, so we worked through two batches and therefore had to prepare another batch of driselase. Our handling had improved, yet it still took twelve hours and again, there was no time left for a break. After we said good bye, we celebrated with a couple of beers and some yummy flammkuchen at the UC uni café of Freiburg. We had learned so much and got much closer to our goal. A big successful step for our team!
| + | |
| - | | + | |
| - | ==4. Regeneration and Selection of transgenic plants==
| + | |
| - | After Transformation, the regenerating protoplasts were incubated in regeneration medium in 6-well-plates sealed with parafilm for 10 days and then cultivated on a layer of autoclaved cellophane (seperated with Whatman paper during autoclavation, so they dont stick together), on top of Knop medium agar plates for three days under standard conditions. Then we transferred the cellophane layers with the regenerated moss onto Knop medium agar plates containing 25 µg/ml G418 antibiotic for two weeks. The official protocol schedules a two week release period followed by another selection period to ensure stabile transormation, but we didn´t have that much time and went with a single round of selection. We plated only half of our transformed protoplasts. The other half was left in the 6-well-plates where 2 ml of selection medium were added to the 2 ml of regeneration medium from the transormation, with G418 diluted 1:8000.
| + | |
| - | | + | |
| - | ==5. Characterization of transgenic plants==
| + | |
| - | | + | |
| - | ==5.1 Testing our modular moss receptor==
| + | |
| - | ===Idea - GFP fluorescence===
| + | |
| - | Already during the planning phase of the trans membrane receptor we thought about how to track the localization of the receptor – or in other words how to proof the correct folding into the plasma membrane. As a very simple tool we fused genetically a GFP molecule C-terminal to the trans membrane domain of the receptor. A green fluorescence localized at the plasma membrane would hence indicate a correct folding of our receptor into the plasma membrane.
| + | |
| - | [[File:TUM13_FLUA_MEMB.gif|thumb|center|920px|'''Figure 5:''' Z-stack of transgenic moss cells expressing the transmembrane receptor ]]
| + | |
| - | The animated Z-stack of transgenic moss cells expressing the trans membrane receptor presented in Figure 5 shows a bright green fluorescence as is characteristic for GFP. This fluorescence is mainly located at the plasma membrane which would indicate that our receptor proteins have the correct localization. In comparison to the Z-stack taken from transgenic moss expressing GFP in the cytoplasm which is shown in figure six the fluorescence of the moss carrying the trans membrane receptor seems to be more associated to the plasma membrane. However voluminous vacuoles can compress the cytoplasm to a thin layer so that the green fluorescence of cytoplasmic proteins would appear as membrane associated. Hence the fluorescence shown in figure 5 is a first indication that our receptor works properly but cannot serve as a reliable evidence. Therefore we decided to execute a second method for the proof of localization of our trans membrane receptor. Furthermore it is planned for the future to do confocal laser microscopy using fluorescein-biotin conjugate and quantum dots coupled to streptavidin. Therewith the localization of the trans membrane receptor carrying FluA as an effector domain at the N-terminus should be proofed doubtless through bright fluorescence of the used fluorophores on the outside of the plasma membrane and green fluorescence of the receptor GFP on the inner side of the plasma membrane.
| + | |
| - | <html><div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:514px;"><a href="/File:TUM13_GFP_CYT.gif" class="image"><img alt="" src="https://static.igem.org/mediawiki/2013/0/03/TUM13_GFP_CYT.gif" width="512" height="512" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:TUM13_GFP_CYT.gif" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div><b>Figure 6:</b> Z-stack of transgenic moss cells expressing GFP in the cytoplasm</div></div></div></div></html>
| + | |
| - | | + | |
| - | ===2. Idea - Protease cleavage of the effector protein by TEV protease===
| + | |
| - | [[File:TUM13_SERK-Receptor.png|thumb|right|250px|'''Figure 7:''' Schematic design of an effector, in this context effector = Nanoluciferase, immobilised on a transmembrane domain.]]
| + | |
| - | We want to investigate the localisation of our synthetic receptor. For this purpose we used a transgenic moss strain which contains NanoLuc luciferase at the extracellular domain of the receptor. Between the luciferase and the transmembrane region we have added a linker consisting of a ''Strep''tag II and a TEV cleavage site. By the addition of recombinant TEV protease, the TEV protease should be able to pass the cell wall and enter the appoplast, cleave the TEV cleavage site and to liberate the NanoLuc luciferase. This luciferase can than be analyzed by its luminescense, the ''Strep''tag or by mass spectrometry.
| + | |
| - | | + | |
| - | To prove this process, we cut off our Nanoluciferase-Streptag-Construct by using the TEV-Protease. As we hypothetically know, our membrane construct is localized in the membrane and the Nanoluciferase-Streptag-Construct orientated to the Periplasma, protected by the cell-wall of our moss. In conclusion to that, the TEV protease needs to pass the cell-wall to cut off the Nanoluciferase-Streptag-Construct. This process is possible in liquid medium, because the TEV protease just has a molecular weight between 25 kDa and 27 kDa, so it can pass the cell-wall smoothly. After the TEV-Protease has entered the Periplasma of our moss cells, it cuts off the Nanoluciferase-Streptag-Construct at the TEV cleavage site. Again, this construct just has in sum a molecular weight of 20 kDa (Nanoluciferase - 19 kDa, Streptag - 1 kDa), so it can pass the cell-wall due to diffusion to get to the exterior of the moss cell.
| + | |
| - | | + | |
| - | After this experimental part, it is necessary to prove the theoretical deliberation. In the following we performed several experiments like SDS-Pages, Luciferase Assays and tried to bind our construct by utilizing different beads.
| + | |
| - | | + | |
| - | ===Experimental Setup for localization process of membrane bound Nanoluciferase===
| + | |
| - | | + | |
| - | '''First Step'''
| + | |
| - | | + | |
| - | The reagents which were necessary to perform the first step have been 1,9 ml TEV Protease Buffer, 100 µl TEV Protease and as much genetically modified moss, containing the membrane bound Streptag-Luciferase-Construct, as possible. All reagents have been added to a 2 ml Tube, gently mixed and incubated over night at 4°C.
| + | |
| - | | + | |
| - | '''Second Step'''
| + | |
| - | | + | |
| - | In the second step, the 2 ml tube was centrifuged at 13200 rpm for 5 minutes to make the moss cells form a pellet at the bottom of the tube. Afterwards the supernatant was transferred to a new 2 ml tube to isolate the supernatant where the Nanoluciferase-Streptag-Construct was expected to be found. Two slightly different approaches of Immunoprecipitation have been performed to try to bind the cut-off construct.
| + | |
| - | | + | |
| - | First 1 ml of the supernatant was incubated with 20 µl of an antibody called Strepmab-Immo (1 mg/ml), which is able to bind the cut-off construct at the Streptag. After an incubation time of four hours, 100 µl Dynabeads spiked with Protein G have been added to the tube and incubated over night. Protein G is able to bind the Fc-Part of the Strepmab-Immo Antibody. The aim is to bind the Streptag-Nanoluciferase-Construct with the Strepmab-Immo Antibody and the resulting construct with the Protein G anchored magnetic Dynabeads. To get to know if the resulting construct of Protein-G Dynabeads and Strepmab-Immo Antibody are fluorescent active in the same range of the electromagnetic spectrum like the Nanoluciferase, a negative control containing 5 µl Dynabeads and 1 µl Strepmab-Immo Antibody was prepared.
| + | |
| - | | + | |
| - | The second approach was to use Biotin-labeled beads to bind the Nanoluciferase-Streptag-Construct. Again 20 µl of Biotin labeled beads have been added to 1 ml of the supernatant, carefully mixed and incubated over night. A negative control containing Phosphate-Bufferd-Saline (PBS) buffer and 5 µl Biotin-labeled beads was established, because of the same reason described above, too.
| + | |
| - | | + | |
| - | '''Third step'''
| + | |
| - | | + | |
| - | In the third step both approaches have been centrifuged at 13200 rpm for two minutes to discard the beads. Each supernatant was transferred to a separate 2 ml tube, so we could measure if the beads have bound the antibody-nanoluciferase-streptag and nanoluciferase-streptag construct by comparing the supernatants with the samples containing the beads. The beads have been washed three times with 2 ml of PBS-Buffer. After measuring the Luciferase activity in each of the four samples in a 96 well plate with 20 µl of each sample mixed with 80 µl of Luciferase-Substrate-Buffer using a plate reader, we clamped that the Luciferase activity in the supernatant was at its limit, whereas every bead-sample showed no significant Luciferase activity.
| + | |
| - | | + | |
| - | '''Fourth step'''
| + | |
| - | | + | |
| - | The conclusion of step three was that the beads have not been able to bind the cut-off construct, but, because of the significant Luciferase activity in both supernatants, it stands to reason that the membrane construct was expressed by the moss in a significant way. To get to know more about the cut-off construct we started concentrating cut-off construct in the supernatants to a volume of 150 µl.
| + | |
| - | | + | |
| - | '''Fifth step'''
| + | |
| - | | + | |
| - | After the cut-off construct in the supernatants was concentrated, we performed a SDS-Page with three samples to get to know how big the cut-off construct was. Each sample was prepared by mixing 15 µl supernatant with 7 µl of Sodiumdodecylsulfat and cooked at 95°C. Afterwards, the samples have been applied to a standard SDS-Gel and unseamed. The result of the SDS-Page '''proves the existence''' of the cut-off Nanoluciferase-Streptag-Construct at about 20 kDa. The second band at about 150 kDa refers to the used Antibody in step 1.
| + | |
| - | | + | |
| - | '''Sixth step'''
| + | |
| - | | + | |
| - | A standard Western Blot was performed, using two different Antibodies to label the cut-off Luciferase-Streptag-construct. First we labeled the cut-off construct with Strepmab Classic. Afterwards we labeled the Strepmab Classic with an Antibody fused to the enzyme Alkaline Phosphatase to bind the Strepmab Classic Antibody. By starting the reaction process of the Alkaline Phosphatase using NBP and BCIP as substrate and AP-Buffer, we tried to make the cut-off construct visible on the PVDF membrane. On the Western Blot, no significant band was seen.
| + | |
| - | | + | |
| - | '''Conclusion'''
| + | |
| - | | + | |
| - | During the whole experiment, three different approaches to try to label the Streptag of the cut-off construct, did not work as expected. The easiest explanation of this outcome would be that there has not been formed any cut-off construct. In contrast to that, the Luciferase-Assay showed a significant activity rate, so there has to be a significant amount cut-off Luciferase in the supernatant. Further on the SDS page showed two bands. The first at about 150 kDa belonging to the Antibody used during the DynaBead approach whereas the second band was seen at about 20 kDa belonging to the cut-off Nanoluciferase-Streptag-Construct. <br>
| + | |
| - | An explanation for these results could be that the Streptag, fused to the Nanoluciferase, has not been accessible for any Antibody engineered against Streptag. Due to the seen band at 20 kDa on the SDS page and the significant luciferase activity in the luciferase assay, '''it is experimentally proven''' that our membrane construct is expressed and stocked in the moss's cell membrane.
| + | |
| - | | + | |
| - | ==5.2 Comparison of Signal Peptides for Secretion of Effector Proteins==
| + | |
| - | [[File:TUM13_Result_SigP_comparison.png|thumb|right|500px|'''Figure 8''': Work flow for the generation of transgenic moss.]]
| + | |
| - | In our project the differential localisation of effector proteins was an important task which is also important for later iGEM teams who are interested in working with ''Physcomitrella patens''. Through out the literature there are several different signal peptides which have been used to secrete proteins out of the moss cell. We decided to use two different signal peptides in our project. The first one is the Igkappa secretion signal from ''Mus musculus'' which has been recently shown to be functional as a signal peptide in ''Physco'' [http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009]. Beside this proven signal peptide we also had the signal peptide which we derived from the SERK receptor. As it was interesting for us to know which of these signal peptides workes best we cloned both of them ahead of the NanoLuc luciferase in RFC[25]. As you can see above we cloned these genes of interest into our expression vector for stable integration. After the transfection we selected for four weeks stable transgenic moss lines using the antibiotic G418. In the end we picked 8 clones from the corresponding plates and performed an analysis of 8 single clones. This is necessary as the localisation of the transgene within the gene can have drastic effects on the expression strength of the transgene.<br>
| + | |
| - | | + | |
| - | For the experiments 100 µl of Knop-media at pH 6.5 were transferred into 96-well plates. Subsequently single moss colonies were added to these wells and the the moss plants were incubated for 12 h in order to give the plants enough time to secrete the NanoLuc luciferase with the different signal peptides. The next day 50 µl of the supernatant in which the cells were cultivated was transfered to an empty well. The NanoGlow substrate (sponsored by Promega) was diluted in Water and 50 µl of the substrate was added rapidly to the 96-well plate. The luminescense was quantified for 1 sec in a BioTek II plate reader with a filter of 460 nm.
| + | |
| - | | + | |
| - | '''Results: '''The comparison of the two signal peptides gave a very clear result in our experiment. The Igkappa signal sequence only gives a luminescense signal which is slightly higher than the signal obtained for the wild type plants for which the autoluminescense of the substrate is responsible for the signal. In contrast the SERK signal peptide yielded luminescense signals in the supernatant that were about 1000-fold higher compared to the Igkappa signal sequence. This is also the proof that we successfully integrated a device encoding a secreted luciferase which is efficiently secreted by the moss into the supernatant. This is one of the major obstacles we had to solve in order to be able to secrete effector proteins by our PhyscoFilter.
| + | |
| - | | + | |
| - | ==5.3 The PhyscoFilter for Erythromycin==
| + | |
| | In order to check the activity of the erythromycine esterase B we pursued two different approaches: on the one hand we produced the enzyme recombinant in ''Escherichia coli'' and purified it. The purified protein was used for in vitro activity tests determining the depletion rates. The reaction was stopped at predetermined times by addition of three volumes acetonitrile and freezing in liquid nitrogen. On the other hand we added the substrate erythromycin at a concentration of 0.2 g L<sup>-1</sup> to 3 mL of a polyclonal suspension of transgenic moss plants expressing the erythromycin esterase B in the cytoplasm. After 24 h of incubation at normal growth conditions 100 µL of the supernatant were used for further analysis. | | In order to check the activity of the erythromycine esterase B we pursued two different approaches: on the one hand we produced the enzyme recombinant in ''Escherichia coli'' and purified it. The purified protein was used for in vitro activity tests determining the depletion rates. The reaction was stopped at predetermined times by addition of three volumes acetonitrile and freezing in liquid nitrogen. On the other hand we added the substrate erythromycin at a concentration of 0.2 g L<sup>-1</sup> to 3 mL of a polyclonal suspension of transgenic moss plants expressing the erythromycin esterase B in the cytoplasm. After 24 h of incubation at normal growth conditions 100 µL of the supernatant were used for further analysis. |
| | | | |
| | Both samples were analyzed by liquid chromatography coupled to mass spectrometry (UPLC/MS/MS). The samples were measured at a Waters Xevo TQ-S using a Waters Acquity BEH C18 2,1 x 100mm column (1.7 µm, 130 Å pore size) and the following method: Solvent A: 0.1 % formic acid in water, B: 0.1 % formic acid in ACN and a linear gradient of 99 % A 0.5 min isocratic to 99 % B in 6 min. Column temperature 40 °C, flux 0.4 mL min<sup>-1</sup>, maximal pressure 703 bar. The tuning was executed with solutions consisting of the pure educt at concentrations of approximately 0.1 mgL<sup>-1</sup>. | | Both samples were analyzed by liquid chromatography coupled to mass spectrometry (UPLC/MS/MS). The samples were measured at a Waters Xevo TQ-S using a Waters Acquity BEH C18 2,1 x 100mm column (1.7 µm, 130 Å pore size) and the following method: Solvent A: 0.1 % formic acid in water, B: 0.1 % formic acid in ACN and a linear gradient of 99 % A 0.5 min isocratic to 99 % B in 6 min. Column temperature 40 °C, flux 0.4 mL min<sup>-1</sup>, maximal pressure 703 bar. The tuning was executed with solutions consisting of the pure educt at concentrations of approximately 0.1 mgL<sup>-1</sup>. |
| | | | |
| - | [[File:TUM13_EreB_LCMS.png|thumb|center|910px|'''Figure 9''': Chromatograms with assoziated masses for the LCMS analysis of (A) pure erythromycine as a standard, (B) erythromycine incubated with purified erythromycine esterase B and (C) erythromycine incubated with transgenic moss expressing erythromycine esterase B in the cytoplasm.]] | + | [[File:TUM13_EreB_LCMS.png|thumb|center|910px|'''Figure 1''': Chromatograms with assoziated masses for the LCMS analysis of (A) pure erythromycine as a standard, (B) erythromycine incubated with purified erythromycine esterase B and (C) erythromycine incubated with transgenic moss expressing erythromycine esterase B in the cytoplasm.]] |
| | | | |
| | In part A of figure seven the liquid chromatography chromatogram and the associated molecular mass for pure erythromycin as a standard is shown highlighted in red at a retention time of 3.65 min and a mass of ~734.3 g mol<sup>-1</sup>. | | In part A of figure seven the liquid chromatography chromatogram and the associated molecular mass for pure erythromycin as a standard is shown highlighted in red at a retention time of 3.65 min and a mass of ~734.3 g mol<sup>-1</sup>. |
| Line 290: |
Line 22: |
| | ''' | | ''' |
| | | | |
| - | ==5.4 The PhyscoFilter for Diclofenac and Ethinylestradiol== | + | ==The PhyscoFilter for Diclofenac and Ethinylestradiol== |
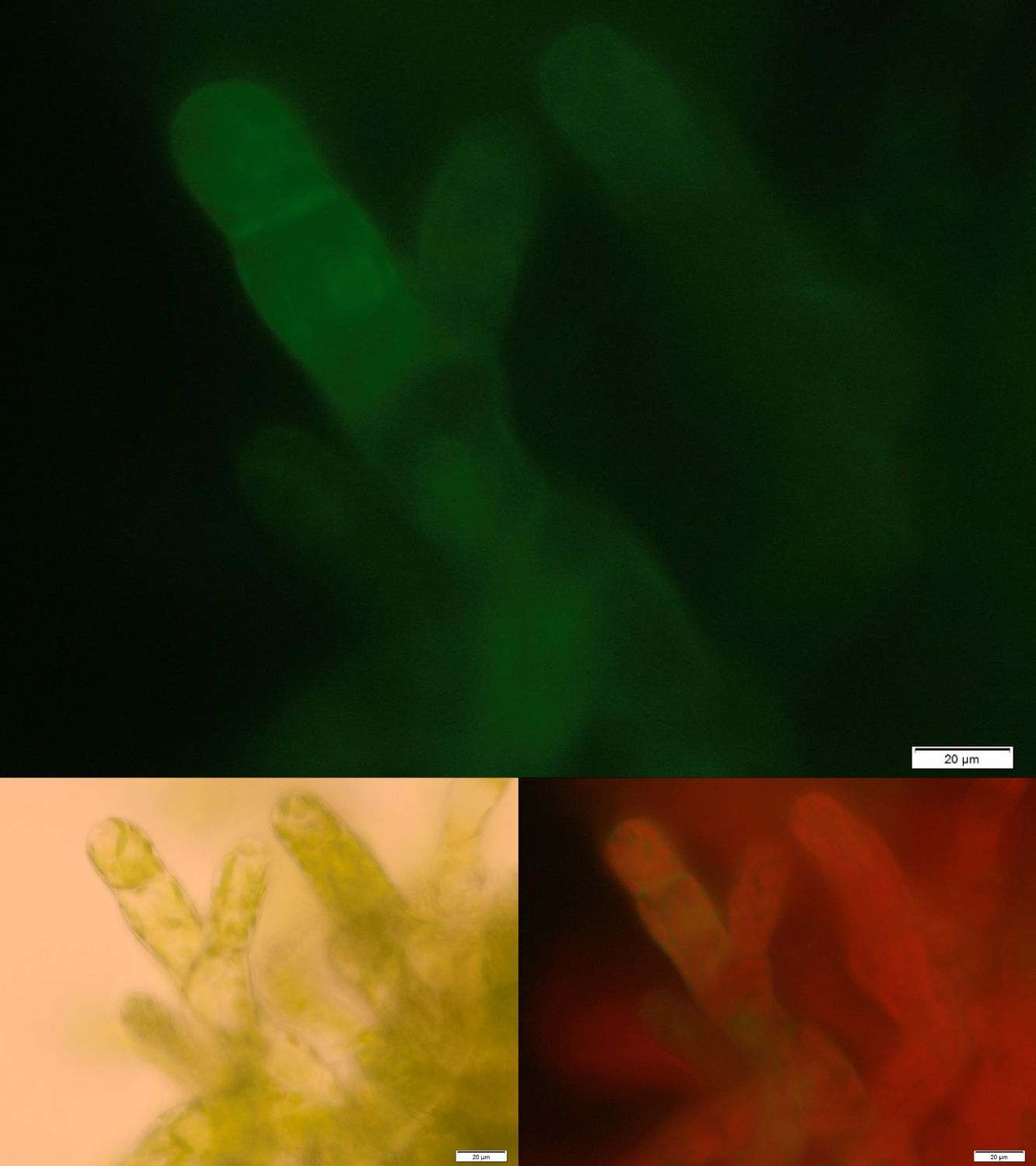
| | + | One major advantage of the applied enzyme laccase is its broad substrate spectrum. The enzyme should be able to degrade important hormones as 17-ß-estradiole or ethinylestradiole as well as the extensively used pain killer diclofenac. Laccases are secretory enzymes requiring an oxidized milieu for proper activity. Therefore we tried to express the laccase from ''Bacillus pumilus'' membrane anchored. |
| | + | [[File:Localization_membrane_Laccase.jpg|thumb|center|500px|'''Figure 2:''' Successfully selected moss with membrane-anchored Laccase. GFP is fused to the intracellular part of the transmembrane region. Upper image: GFP-fluorescence; lower left: brightfield; lower right: GFP-fluorescence plus chloroplast autofluorescence.]] |
| | + | Figure 2 shows fluorescence microscopy images of the receptor associated GFP. In comparison to other fluorescence images of trans membrane constructs the fluorescence at the membrane is ony slightly increased. This could indicate an inefficient integration of our construct into the membrane. Therefore we tried to determine if our transgenic moss is able to degrade substrate ''in vivo''. Besides LC MS a suitable detection method for diclofenac is a competitive ELISA developed by prof. Knopp. This ELISA is by now commonly used for the routine detection of diclofenac in waste water. |
| | + | [[File:TUM13_ELISA_schema.png|thumb|center|920px|'''Figure 3:''' Scheme of the performed competitive ELISA to determine the diclofenac concentration.]] |
| | + | [[File:TUM13_ELISA_standard.png|thumb|left|440px|'''Figure 4:''' Measured standardization for diclofenac.]] |
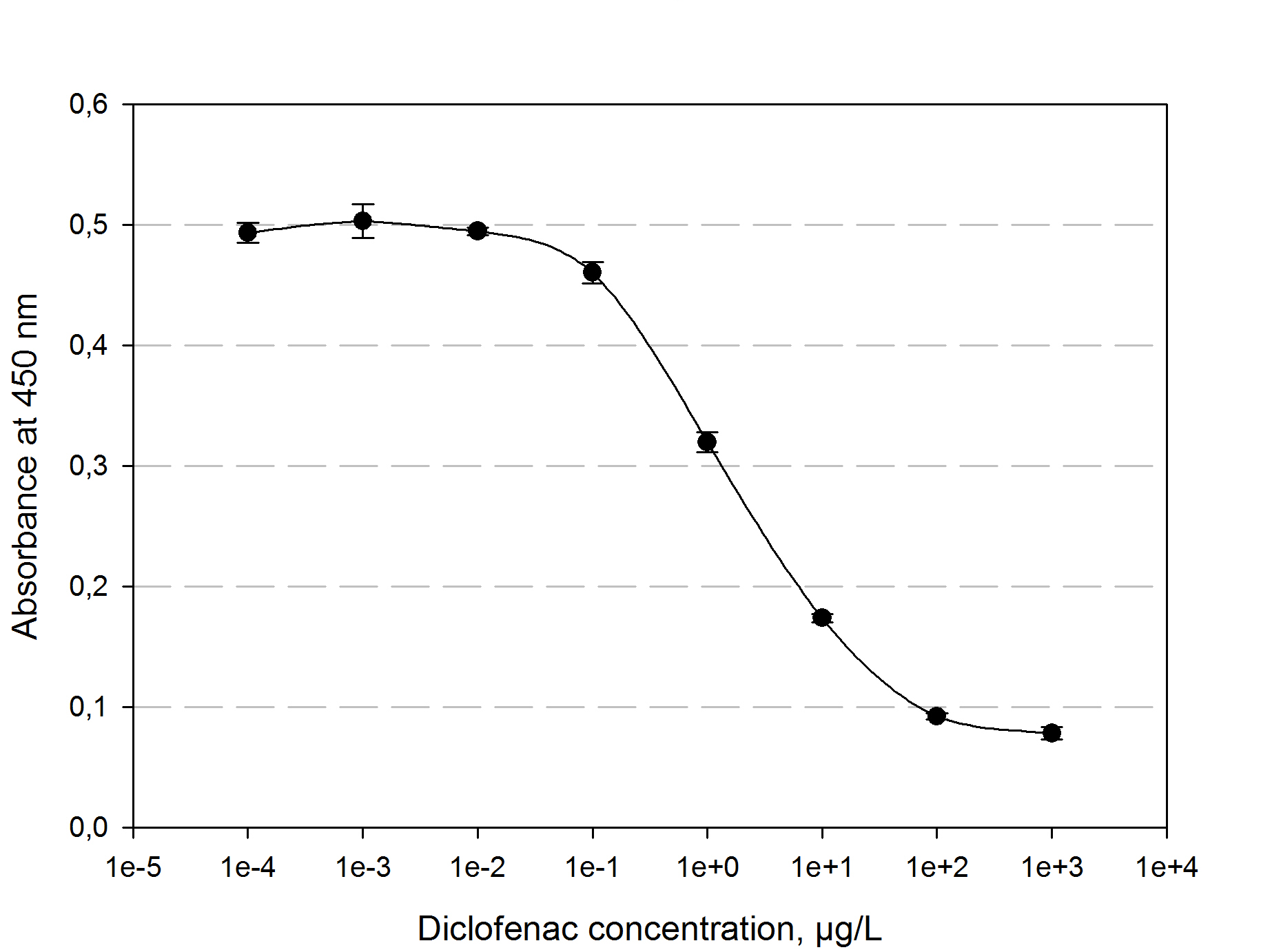
| | + | The function principle of the ELISA is as follows: a 96 well plate gets precoated with an diclofenac-OCA-conjugate at 4°C over night. After washing and blocking with 1% casein the competition reaction is initiated by the addition of a polyclonal antibody solution and the samples which should be quantified. The diclofenac present in the samples competes with the diclofenac-OVA-conjugate to be bound by the antibodies. The more diclofenac a sample contains, the less antibody binds to the plate surface. After a washing step a secondary HRP-labeled antibody is added to the plate. The read-out reaction is initiated by coating the plate with 1% H<sub>2</sub>O<sub>2</sub> and TMB (3,3′,5,5′-Tetramethylbenzidin). After 15-20 min. the reaction is stopped by adding 5 % H<sub>2</sub>SO<sub>4</sub>. The blue TMD with an absorbtion maxima at 650 nm changes to yellow with an absorption at 450 nm. The amount of TMB is detected by measuring the absorbance. The standard calibration graph for diclofenac concentrations of 10<sup>-5</sup> to 10<sup>4</sup> µg/L we performed is shown in figure 4. The samples of diclofenac incubated with either transgenic moss or recombinant produced and purified laccase did not show any evaluable results. Possibly the used primary antibody shows cross reactivity with the created product of the degradation reaction, so that the assay is not suitable to quantify the degradation product. This has to be checked via LC MS. |
| | | | |
| - | ==5.5 The PhyscoFilter for Microcstin (receptor bound BioAccumulation)== | + | ==The PhyscoFilter for Microcystin == |
| | + | Besides our own approaches to prove the opportunity of cell surface immobilized effectors for the bioaccumulation of those pollutants which cannot be degraded enzymatically so far, we took part in a collaboration with this year’s iGEM team from Dundee and expressed their Protein Phosphatase 1 in our moss. Due to the high amount of free cysteine residues of this protein it is necessary to express it as a secreted or cell surface immobilized protein. For a sustainable removal of PP1’s substrate microcystein we decided to express PP1 membrane-anchored. The naturally occurring membrane turn-over could lead to a degradation of the receptor-bound microcystein through the intra cellular degradation machinery. This way the idea of a self-regenerating biological filter could be realized for non-enzymatically degradable substances, too. |
| | | | |
| - | ==The PhyscoFilter for Fluorescein (receptor bound BioAccumulation using <partinfo>BBa_K1159003</partinfo>)== | + | [[File:Localization_membrane_PP1.jpg|thumb|center|920px|'''Figure 5:''' Successfully selected moss with membrane-anchored Protein Phosphatase 1. BUT GFP is mainly localized in aggregates in the cytosolic or endoplasmatic environment. Upper images: GFP-fluorescence; lower images: in each case on the left: brightfield and on the right: GFP-fluorescence plus chloroplast autofluorescence.]] |
| | + | |
| | + | In the upper part of Figure 5 fluorescence images of the transgenic moss transformed with membrane-bound PP1 are shown. Conspicuously the fluorescence only occurs in small locally restricted dots which are widespread distributed over the whole cell. Anyway the images give strong indications that the receptor fusion proteins are not membrane associated due to aggregation, assumedly in the endoplasmic reticulum. Probably even the protein folding machinery of our moss cannot handle the correct folding of this construct. Therefore from our point of view protein engineering with subsequent deletion of free cysteine residues could be a suitable next step to decrease the aggregation tendency and thereby increase the amount of membrane localized PP1 with correct folding. |
| | + | |
| | + | ==The PhyscoFilter for Fluorescein == |
| | + | |
| | + | [[File:Localization_membrane_FluA.jpg|thumb|center|920px|'''Figure 6:''' Successfully selected moss with membrane-anchored FluA. You can clearly see the GFP fluorescence of the membrane-anchored FluA in the edges of the moss cells. Upper images: GFP-fluorescence; lower two left: brightfield; lower two right: GFP-fluorescence plus chloroplast autofluorescence.]] |
| | + | |
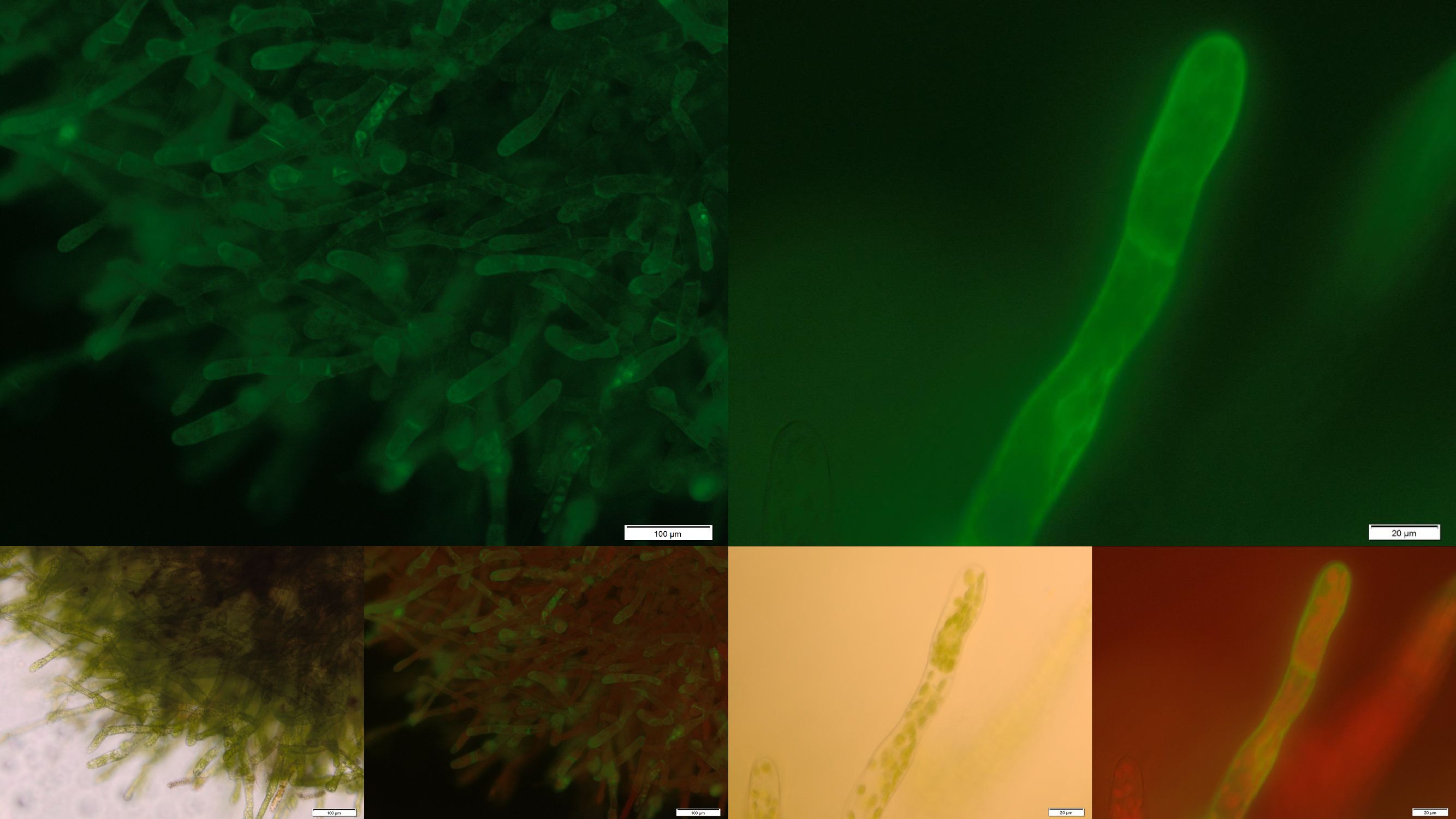
| | + | In order to prove the suitability of surface immobilized binding molecules in general and of aniticalins in particular we decided to transform our moss with a genetic construct containing [http://parts.igem.org/Part:BBa_K1159003 FluA], an Anticalin with a high affinity towards fluorescein, fused at the C-terminus to the N-terminus of our developed [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1159315 trans membrane receptor]. Especially in the upper and lower fluorescence images on the right a compared to the cytoplasm increased membrane associated GFP fluorescence can be seen. This fluorescence indicates that our used secretion signal and trans membrane domain work properly and lead to the desired membrane localization. |
| | | | |
| | ==References:== | | ==References:== |
| Line 301: |
Line 48: |
| | <!-- Ab hier richtige Referenzen einfügen --> | | <!-- Ab hier richtige Referenzen einfügen --> |
| | | | |
| - | #[[http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf]] Hohe, A., T. Egener, J. Lucht, H. Holtorf, C. Reinhard, G. Schween, R. Reski (2004): An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene knockouts in a moss, Physcomitrella patens. Current Genet. 44, 339-347.
| + | [[http://www.plant-biotech.net/paper/CurrGenet_2003_hohe.pdf Hohe et al., 2004]] An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene knockouts in a moss, Physcomitrella patens, Current Genet. 44, 339-347. |
| | + | |
| | + | [[http://www.ncbi.nlm.nih.gov/pubmed/12966990 Deng et al., 2003]] Residue analysis of the pharmaceutical diclofenac in different water types using ELISA and GC-MS, Environ Sci Technol. 37, 3422-3429. |
| | | | |
| | <!-- End of content --> | | <!-- End of content --> |

 "
"





AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351