Team:TU-Munich/Results/Localization
From 2013.igem.org
(→Experimental Setup) |
(→SpyCatcher & SpyTag) |
||
| (45 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
<!-- Start of content --> | <!-- Start of content --> | ||
| - | == | + | ==Localization== |
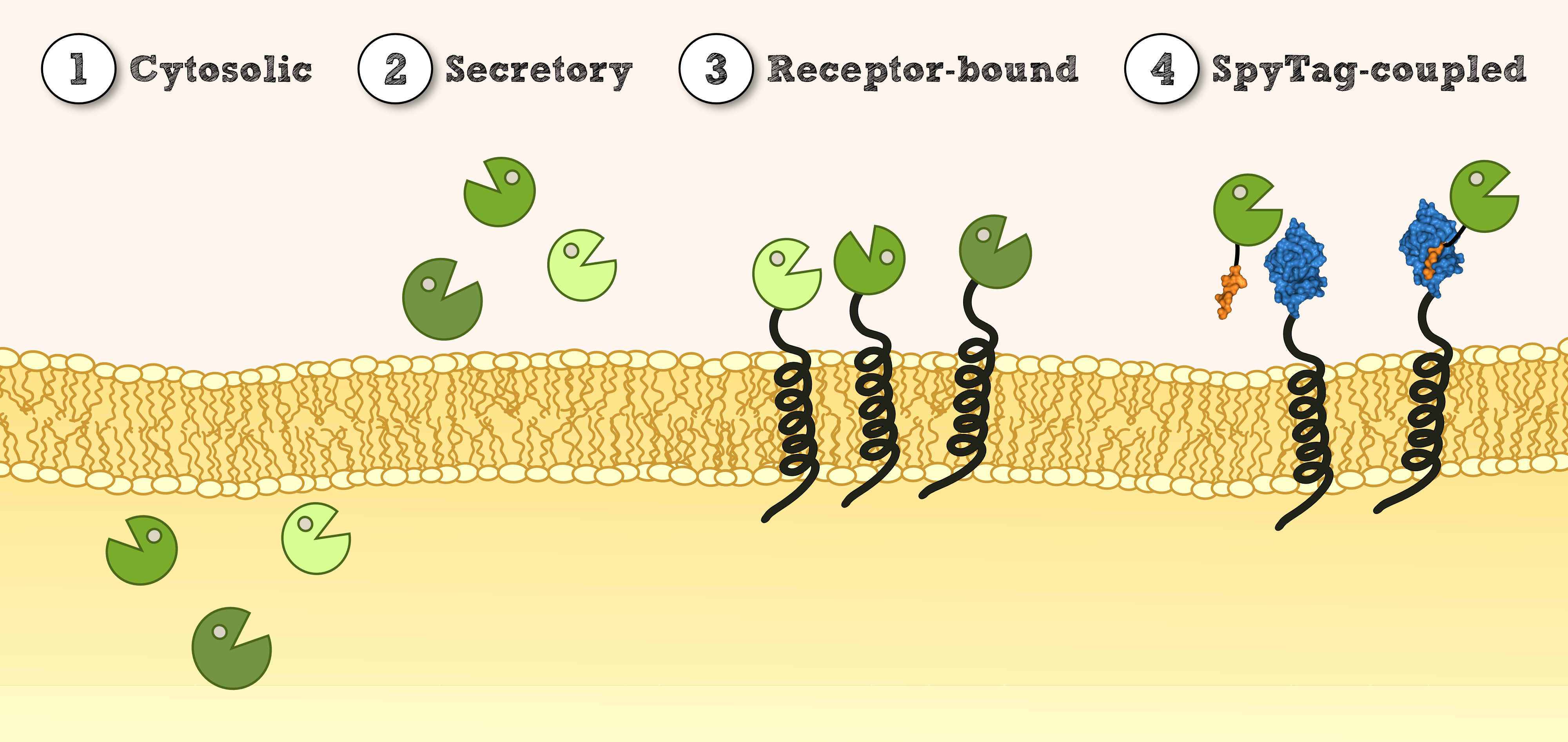
| - | === Experimental Setup === | + | [[File:TUM13_Lokalisation_1.4.png|thumb|center|910px| '''Figure 1''': Overview of different possible localisations for effector proteins.]] |
| - | In our project the differential localisation of effector proteins was an important task which is also important for later iGEM teams who are interested in working with ''Physcomitrella patens''. Through out the literature there are several different signal peptides which have been used to secrete proteins out of the moss cell. We decided to use two different signal peptides in our project. The first one is the Igkappa secretion signal from ''Mus musculus'' which has been recently shown to be functional as a signal peptide in ''Physco'' [http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009]. Beside this proven signal peptide we also had the signal peptide which we derived from the SERK receptor. As it was interesting for us to know which of these signal peptides workes best we cloned both of them ahead of the NanoLuc luciferase in RFC[25]. As you can see above we cloned these genes of interest into our expression vector for stable integration. After the transfection we selected for four weeks stable transgenic moss lines using the antibiotic G418. In the end we picked 8 clones from the corresponding plates and performed an analysis of 8 single clones. This is necessary as the localisation of the transgene within the gene can have drastic effects on the expression strength of the transgene. | + | |
| + | === Secretion via SERK signal peptide (SERK-SigP) === | ||
| + | |||
| + | ==== Experimental Setup ==== | ||
| + | In our project the differential localisation of effector proteins was an important task which is also important for later iGEM teams who are interested in working with ''Physcomitrella patens''. Through out the literature there are several different signal peptides which have been used to secrete proteins out of the moss cell. We decided to use two different signal peptides in our project. The first one is the Igkappa secretion signal from ''Mus musculus'' which has been recently shown to be functional as a signal peptide in ''Physco'' [[http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009]]. Beside this proven signal peptide we also had the signal peptide which we derived from the SERK receptor. As it was interesting for us to know which of these signal peptides workes best we cloned both of them ahead of the NanoLuc luciferase in RFC[25]. As you can see above we cloned these genes of interest into our expression vector for stable integration. After the transfection we selected for four weeks stable transgenic moss lines using the antibiotic G418. In the end we picked 8 clones from the corresponding plates and performed an analysis of 8 single clones. This is necessary as the localisation of the transgene within the gene can have drastic effects on the expression strength of the transgene. | ||
| - | [[File:TUM13_Result_SigP_comparison.png|thumb|right|500px|'''Figure | + | [[File:TUM13_Result_SigP_comparison.png|thumb|right|500px|'''Figure 2''': Work flow for the generation of transgenic moss.]] |
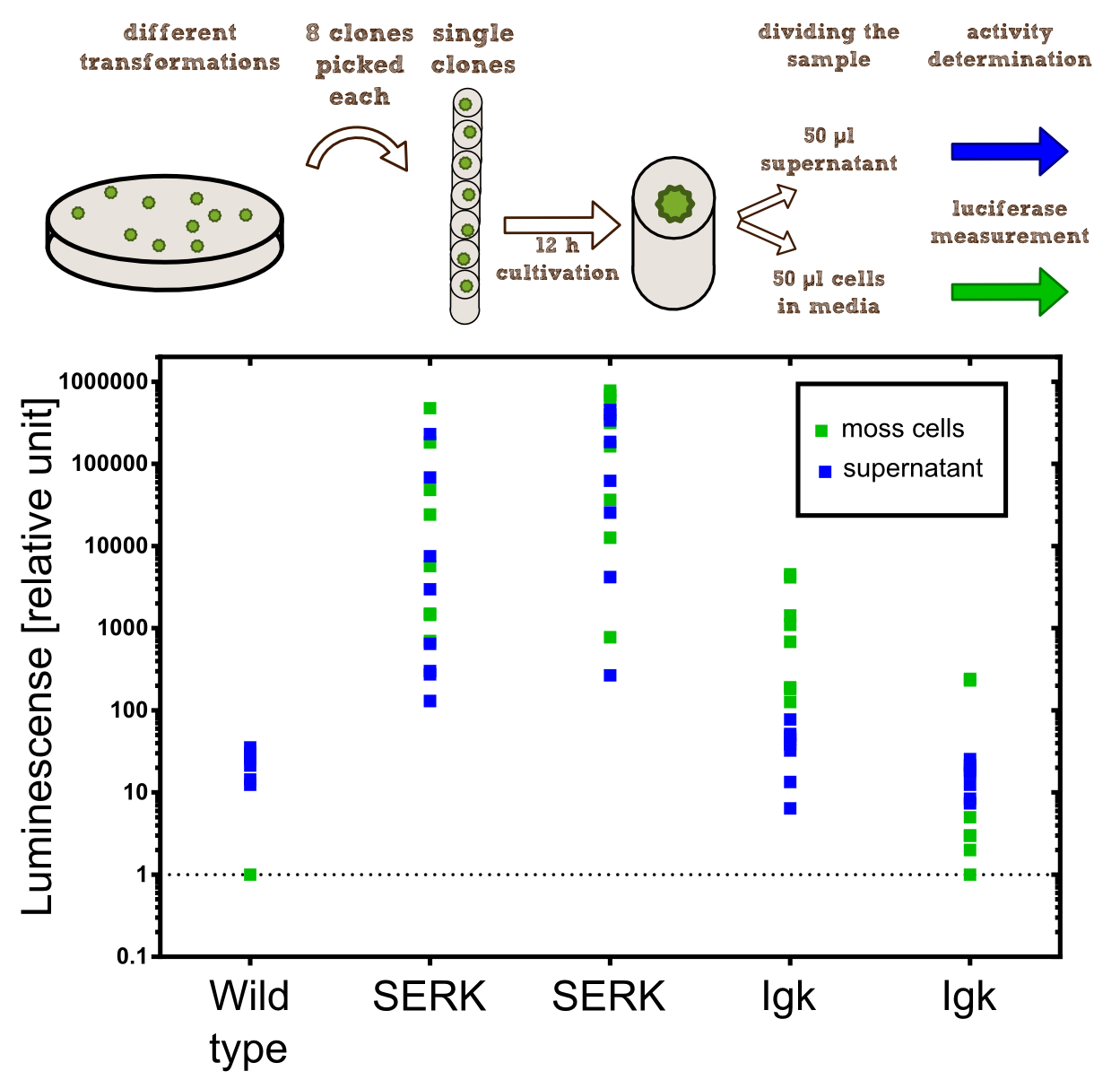
For the experiments 100 µl of Knop-media at pH 6.5 were transferred into 96-well plates. Subsequently single moss colonies were added to these wells and the the moss plants were incubated for 12 h in order to give the plants enough time to secrete the NanoLuc luciferase with the different signal peptides. The next day 50 µl of the supernatant in which the cells were cultivated was transfered to an empty well. The NanoGlow substrate (sponsored by Promega) was diluted in Water and 50 µl of the substrate was added rapidly to the 96-well plate. The luminescense was quantified for 1 sec in a BioTek II plate reader with a filter of 460 nm. | For the experiments 100 µl of Knop-media at pH 6.5 were transferred into 96-well plates. Subsequently single moss colonies were added to these wells and the the moss plants were incubated for 12 h in order to give the plants enough time to secrete the NanoLuc luciferase with the different signal peptides. The next day 50 µl of the supernatant in which the cells were cultivated was transfered to an empty well. The NanoGlow substrate (sponsored by Promega) was diluted in Water and 50 µl of the substrate was added rapidly to the 96-well plate. The luminescense was quantified for 1 sec in a BioTek II plate reader with a filter of 460 nm. | ||
| - | === Results === | + | ==== Results ==== |
The comparison of the two signal peptides gave a very clear result in our experiment. The Igkappa signal sequence only gives a luminescense signal which is slightly higher than the signal obtained for the wild type plants for which the autoluminescense of the substrate is responsible for the signal. In contrast the SERK signal peptide yielded luminescense signals in the supernatant that were about 1000-fold higher compared to the Igkappa signal sequence. This is also the proof that we successfully integrated a device encoding a secreted luciferase which is efficiently secreted by the moss into the supernatant. This is one of the major obstacles we had to solve in order to be able to secrete effector proteins by our PhyscoFilter. | The comparison of the two signal peptides gave a very clear result in our experiment. The Igkappa signal sequence only gives a luminescense signal which is slightly higher than the signal obtained for the wild type plants for which the autoluminescense of the substrate is responsible for the signal. In contrast the SERK signal peptide yielded luminescense signals in the supernatant that were about 1000-fold higher compared to the Igkappa signal sequence. This is also the proof that we successfully integrated a device encoding a secreted luciferase which is efficiently secreted by the moss into the supernatant. This is one of the major obstacles we had to solve in order to be able to secrete effector proteins by our PhyscoFilter. | ||
| - | == Membrane-anchor via SERK transmembrane domain (SERK-TMD) == | + | === Membrane-anchor via SERK transmembrane domain (SERK-TMD) === |
| - | + | ====Verification of membrane integration by epifluorescence microscopy==== | |
| - | + | [[File:TUM13_SERK-Receptor.png|thumb|right|250px|'''Figure 3:''' Schematic design of an effector, in this context effector = Nanoluciferase, immobilised on a transmembrane domain.]] | |
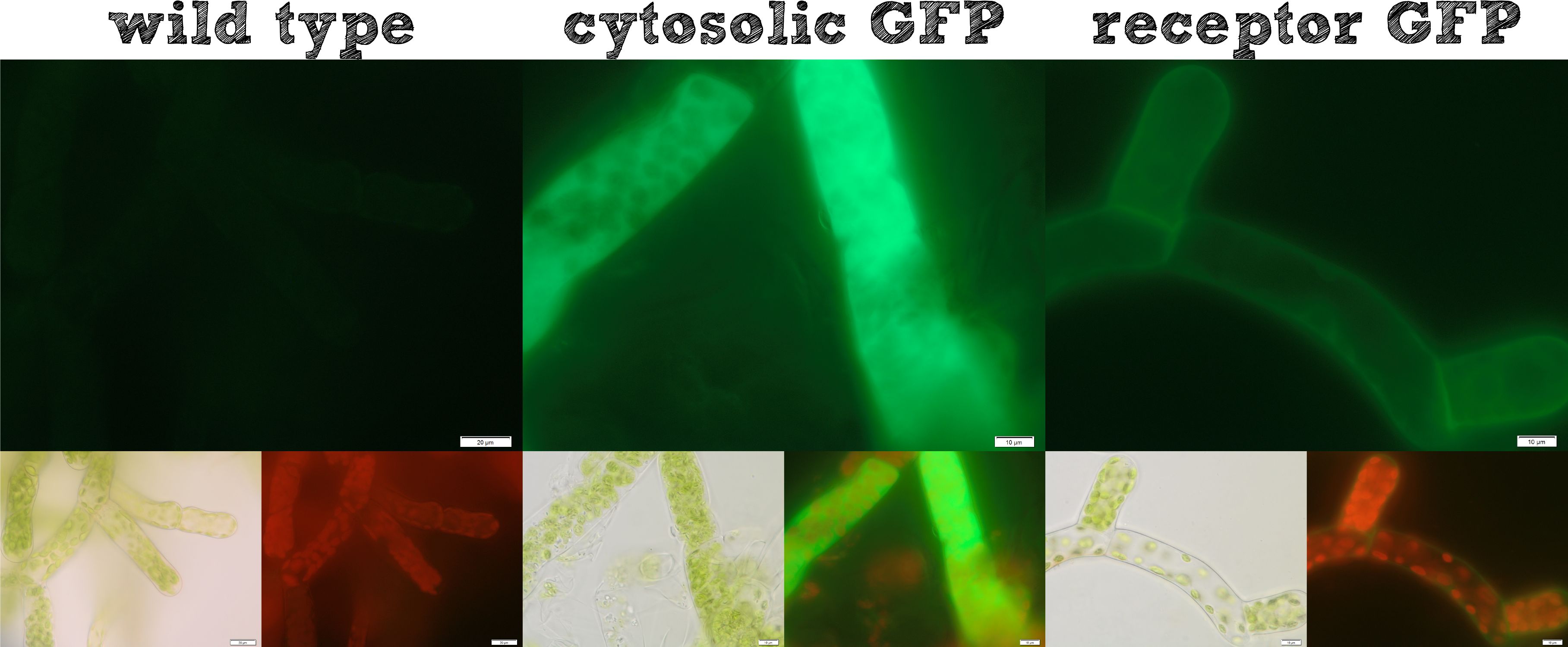
Already during the planning phase of the trans membrane receptor we thought about how to track the localization of the receptor – or in other words how to proof the correct folding into the plasma membrane. As a very simple tool we fused genetically a GFP molecule C-terminal to the trans membrane domain of the receptor. A green fluorescence localized at the plasma membrane would hence indicate a correct folding of our receptor into the plasma membrane. | Already during the planning phase of the trans membrane receptor we thought about how to track the localization of the receptor – or in other words how to proof the correct folding into the plasma membrane. As a very simple tool we fused genetically a GFP molecule C-terminal to the trans membrane domain of the receptor. A green fluorescence localized at the plasma membrane would hence indicate a correct folding of our receptor into the plasma membrane. | ||
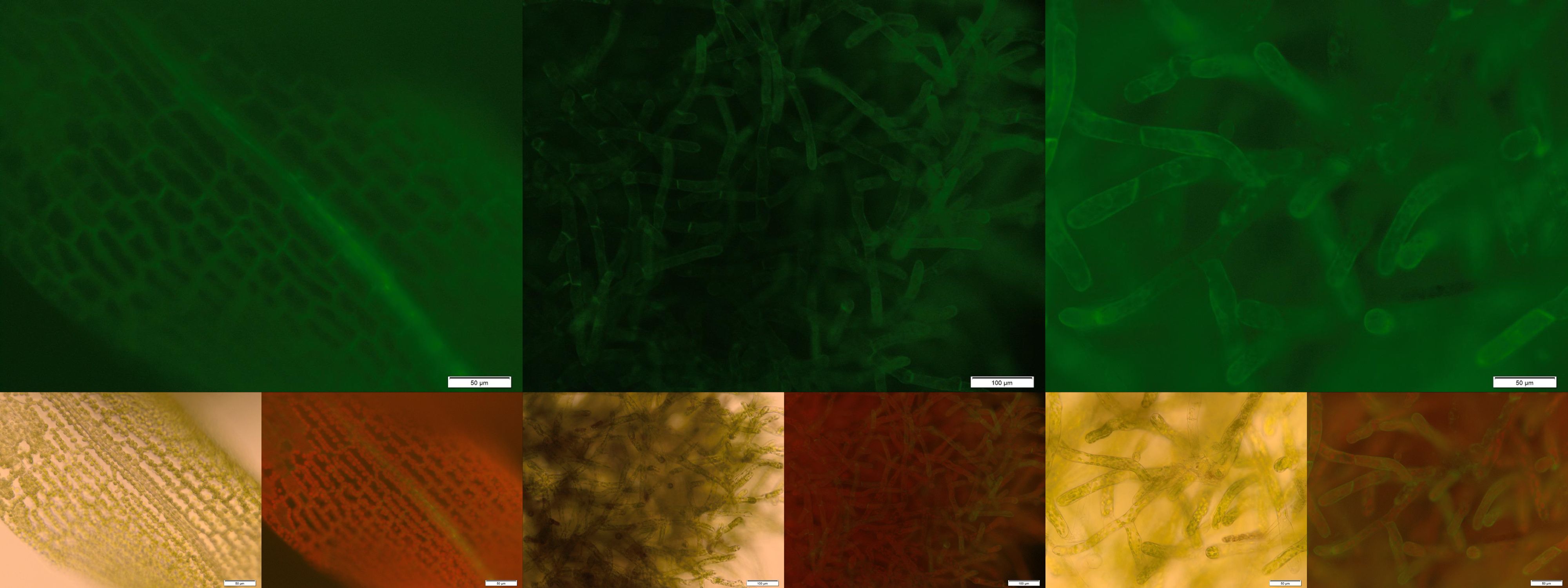
| - | + | The image from epifluorescence microscopy in Figure 4 and 5 shows that the moss cells are expressing the transmembrane receptor presented in Figure 2 shows a bright green fluorescence as is characteristic for GFP. This fluorescence is mainly located at the plasma membrane which would indicate that our receptor proteins have the correct localization. In comparison to the the transgenic moss expressing GFP in the cytoplasm, the fluorescence of the moss carrying the transmembrane receptor seems to be more associated to the plasma membrane. Hence the fluorescence is a first indication that our receptor works properly but cannot serve as a reliable evidence. Therefore we decided to execute a second method for the proof of localization of our transmembrane receptor. | |
| - | [[File: | + | [[File:Localization_general.jpg|thumb|center|920px|'''Figure 4:''' Comparison of different localizations of GFP for testing purposes.]] |
| - | + | [[File:Localization_membrane_nLuc.jpg|thumb|center|920px|'''Figure 5:''' Membrane-anchored NanoLuc luciferase with GFP fusion on the intracellular site of the TMD. Upper image: GFP-fluorescence; lower left: brightfield; lower right: GFP-fluorescence plus chloroplast autofluorescence.]] | |
| - | ===Verification of | + | ====Verification of receptor orientation by shedding off the ecto-NanoLuc luciferase with TEV protease==== |
| - | + | We want to investigate the localisation of our synthetic receptor. For this purpose we used a transgenic moss strain which contains NanoLuc luciferase at the ecto-domain of the receptor. Between the luciferase and the transmembrane region, we have added a linker consisting of a ''Strep''tag II and a TEV cleavage site. By adding the recombinant TEV protease, the TEV protease should be able to pass the cell wall and enter the appoplast, cleave the TEV cleavage site and cut off the NanoLuc luciferase. Afterwards the luciferase in the supernatant can be analyzed by its luminescense. | |
| - | We want to investigate the localisation of our synthetic receptor. For this purpose we used a transgenic moss strain which contains NanoLuc luciferase at the | + | |
| - | To prove this process, we cut off our | + | To prove this process, we cut off our nLuc-''Strep''-tag II-construct by using the TEV-Protease. As we hypothetically know, our membrane construct is localized in the membrane and the nLuc-''Strep''-tag II-construct orientated to the periplasma, protected by the cell-wall of our moss. In conclusion to that, the TEV protease needs to pass the cell-wall to cut off the nLuc-''Strep''-tag II-construct. This process is possible in liquid medium, because the TEV protease just has a molecular weight between 25 kDa and 27 kDa, so it can pass the cell-wall without hindrance. There, it cuts off the nLuc-''Strep''-tag II-construct at the TEV cleavage site. Again, this construct has in sum a molecular weight of 20 kDa (Nanoluciferase – 19 kDa, Streptag – 1 kDa), so it can pass the cell-wall due to diffusion to get to the exterior of the moss cell. |
After this experimental part, it is necessary to prove the theoretical deliberation. In the following we performed several experiments like SDS-Pages, Luciferase Assays and tried to bind our construct by utilizing different beads. | After this experimental part, it is necessary to prove the theoretical deliberation. In the following we performed several experiments like SDS-Pages, Luciferase Assays and tried to bind our construct by utilizing different beads. | ||
| - | ===Experimental Setup=== | + | ====Experimental Setup==== |
| - | ====First Step==== | + | =====First Step===== |
| - | The reagents which were necessary to perform the first step of the experiment have been 1 | + | The reagents which were necessary to perform the first step of the experiment have been 1.863 ml TEV Protease Buffer, 137 µl TEV Protease (0.364 mg/ml) to get a total enzyme activity of 500 Units in each sample and as much genetically modified moss, containing the membrane bound Streptag-Luciferase-Construct, as possible. All reagents have been added to a 2 ml Tube, gently mixed and incubated for 16 hours at 4°C. A negative control containing wild type moss instead of the transgenic one was prepared, too. Before starting the incubation of the prepared samples for 16 hours, we extracted 100 µl of each one, to enable the comparison of the amount of the cut-off construct at the beginning of the incubation time and the incubation over night. |
| - | ====Second Step==== | + | =====Second Step===== |
| - | In the second step both samples, incubated | + | In the second step both samples, incubated for 16 hours, were centrifuged at 10000 rpm for 5 minutes to make the moss cells form a pellet at the bottom of the tube. Afterwards the supernatants, where the Nanoluciferase-Streptag-Construct was expected to be found, were transferred to new 2 ml tubes. |
| - | ====Third step==== | + | =====Third step===== |
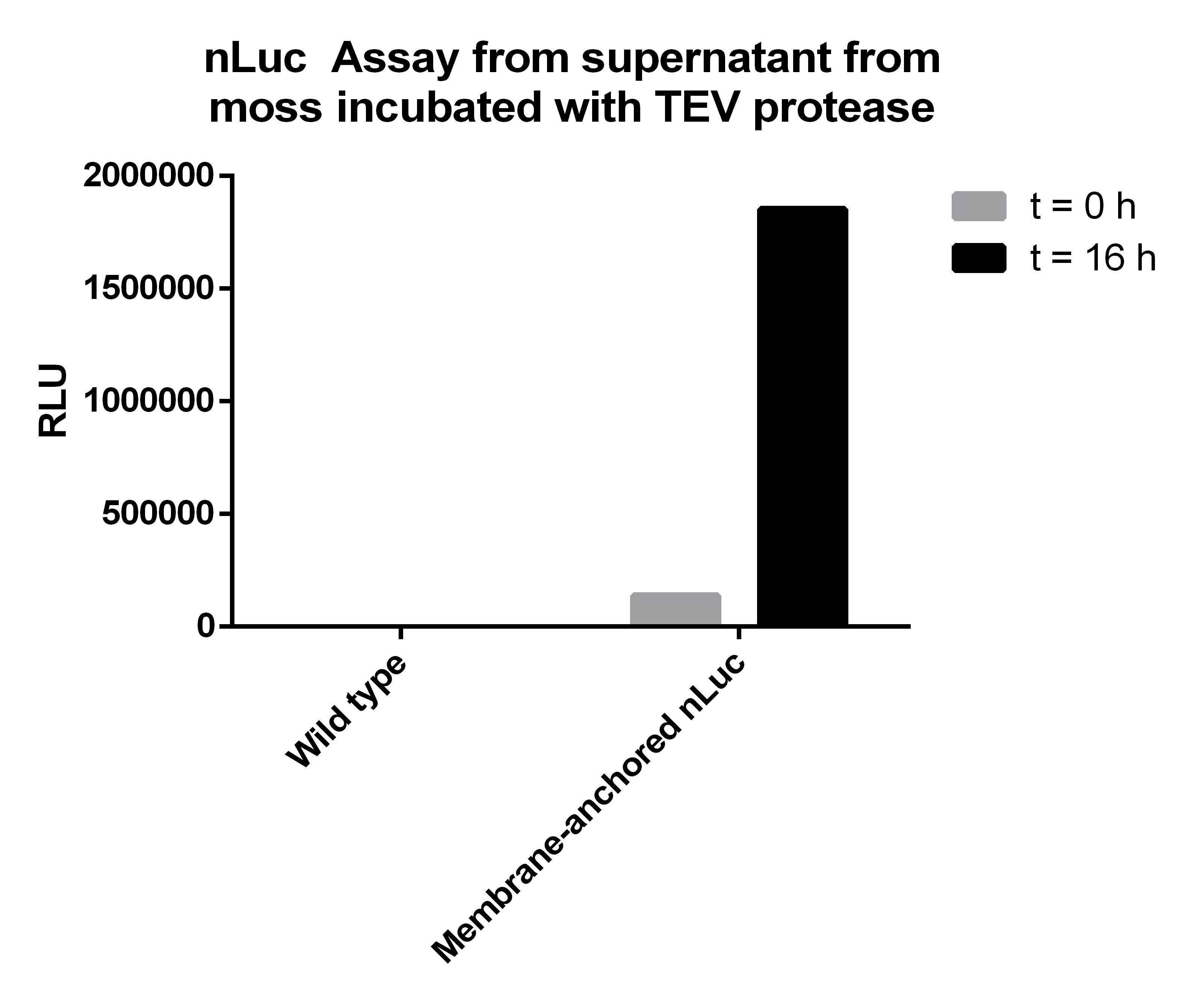
| - | Afterwards we measured the Nanoluciferase activity in the supernatants of the transgenic moss at the beginning of the incubation time and | + | Afterwards we measured the Nanoluciferase activity in the supernatants of the transgenic moss at the beginning of the incubation time and after 16 hours. The same procedure was performed for the Wild-type moss. Additionally we measured the activity of the TEV-Protease buffer in the Luciferase-Assay, to be sure that it does not falsify our results. All samples were prepared with 50 µl supernatant mixed to 50 µl Luciferase-Substrate-Buffer. We concluded that the Luciferase activity was significant in our membrane-construct supernatants, whereas the wild-type moss did not show any significant Luciferase activity. |
| + | |||
| + | [[File:NLucAssay_supernatant_TEV-protease.png|thumb|center|500px|'''Figure 6:''' In the supernatant of moss with membrane-anchored nLuc you can see a dramatic increase of nLuc activity after incubation with TEV protease for 16 hours due to the TEV cleavage site between the nLuc luciferase and the transmembrane region.]] | ||
{|cellspacing="0" border="1" right | {|cellspacing="0" border="1" right | ||
| - | |+ '''Table''' 1: | + | |+ '''Table''' 1: Raw data showed in RLU |
! align="center";| Supernatant transgenic moss - T0 | ! align="center";| Supernatant transgenic moss - T0 | ||
! align="center";| Supernatant transgenic moss - Over night | ! align="center";| Supernatant transgenic moss - Over night | ||
| Line 71: | Line 76: | ||
|} | |} | ||
| - | ====Conclusion==== | + | =====Conclusion===== |
The Luciferase-Assay showed a significant activity rate, so there has to be a significant amount of cut-off Streptag-Nanoluciferase-Construct in the transgenic supernatant. Due to the significant results of the Luciferase-Assay of the transgenic moss supernatant, the '''prove of the existence''' and the desired orientation of our Membrane-Nanoluciferase-Construct is confirmed. | The Luciferase-Assay showed a significant activity rate, so there has to be a significant amount of cut-off Streptag-Nanoluciferase-Construct in the transgenic supernatant. Due to the significant results of the Luciferase-Assay of the transgenic moss supernatant, the '''prove of the existence''' and the desired orientation of our Membrane-Nanoluciferase-Construct is confirmed. | ||
| + | ==SpyCatcher & SpyTag== | ||
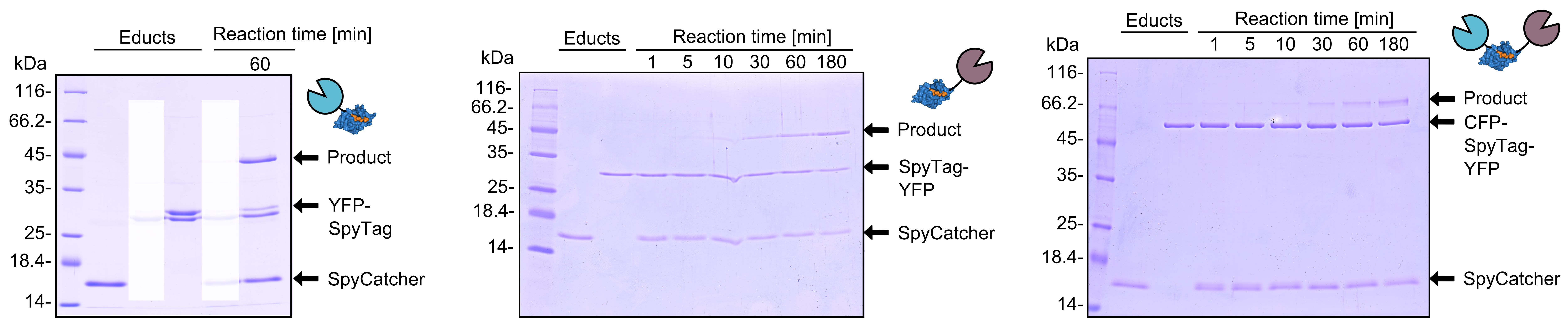
| + | As you can clearly see, all the combinations of orientations of SpyTag with SYFP2 lead to a post-translational covalent isopeptide formation to SpyCatcher (Figure 8). Even fusion to both termini of SpyTag leads to a post-translational fusion to SpyCatcher. So with the SpyCatcher/SpyTag system you can not only post-translationally fuse the C-terminus of the first protein to the N-terminus of another protein; also the system allows post-translational non-canonical protein fusions, like C-C terminal, and branched fusions, which is useful for creating two-step enzyme assembly lines. | ||
| + | |||
| + | [[File:SpySystem_Mechanism_new.png|thumb|right|900px|'''Figure 7. ''' Draft of how our SpyCatcher/SpyTag system works.]] | ||
| + | |||
| + | [[File:TUM13_SpyTag_Exp_4.png|thumb|right|900px|'''Figure 8. '''Reaction of SpyCatcher with SYFP2 fused to the C-terminus and eCFP fused to the N-terminus of SpyTag. Reaction educts were used at a concentration of 2 µM.]] | ||
| + | |||
| + | So by using this system, you can fuse proteins post-translationally in every desired orientation which offers you new oppurtunities in protein engineering. | ||
| + | |||
| + | ==References== | ||
| + | [[http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009]] Functional cross-kingdom conservation of mammalian and moss (Physcomitrella patens) transcription, translation and secretion machineries, Plant Biotechnol J. 2009 Jan;7(1):73-86 | ||
| + | |||
| + | [[http://www.ncbi.nlm.nih.gov/pubmed/22366317 Zakeri et al., 2012]] Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. ''Proc Natl Acad Sci U S A''. 20;109(12)<br> | ||
Latest revision as of 03:57, 29 October 2013
Localization
Secretion via SERK signal peptide (SERK-SigP)
Experimental Setup
In our project the differential localisation of effector proteins was an important task which is also important for later iGEM teams who are interested in working with Physcomitrella patens. Through out the literature there are several different signal peptides which have been used to secrete proteins out of the moss cell. We decided to use two different signal peptides in our project. The first one is the Igkappa secretion signal from Mus musculus which has been recently shown to be functional as a signal peptide in Physco http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009. Beside this proven signal peptide we also had the signal peptide which we derived from the SERK receptor. As it was interesting for us to know which of these signal peptides workes best we cloned both of them ahead of the NanoLuc luciferase in RFC[25]. As you can see above we cloned these genes of interest into our expression vector for stable integration. After the transfection we selected for four weeks stable transgenic moss lines using the antibiotic G418. In the end we picked 8 clones from the corresponding plates and performed an analysis of 8 single clones. This is necessary as the localisation of the transgene within the gene can have drastic effects on the expression strength of the transgene.
For the experiments 100 µl of Knop-media at pH 6.5 were transferred into 96-well plates. Subsequently single moss colonies were added to these wells and the the moss plants were incubated for 12 h in order to give the plants enough time to secrete the NanoLuc luciferase with the different signal peptides. The next day 50 µl of the supernatant in which the cells were cultivated was transfered to an empty well. The NanoGlow substrate (sponsored by Promega) was diluted in Water and 50 µl of the substrate was added rapidly to the 96-well plate. The luminescense was quantified for 1 sec in a BioTek II plate reader with a filter of 460 nm.
Results
The comparison of the two signal peptides gave a very clear result in our experiment. The Igkappa signal sequence only gives a luminescense signal which is slightly higher than the signal obtained for the wild type plants for which the autoluminescense of the substrate is responsible for the signal. In contrast the SERK signal peptide yielded luminescense signals in the supernatant that were about 1000-fold higher compared to the Igkappa signal sequence. This is also the proof that we successfully integrated a device encoding a secreted luciferase which is efficiently secreted by the moss into the supernatant. This is one of the major obstacles we had to solve in order to be able to secrete effector proteins by our PhyscoFilter.
Membrane-anchor via SERK transmembrane domain (SERK-TMD)
Verification of membrane integration by epifluorescence microscopy
Already during the planning phase of the trans membrane receptor we thought about how to track the localization of the receptor – or in other words how to proof the correct folding into the plasma membrane. As a very simple tool we fused genetically a GFP molecule C-terminal to the trans membrane domain of the receptor. A green fluorescence localized at the plasma membrane would hence indicate a correct folding of our receptor into the plasma membrane.
The image from epifluorescence microscopy in Figure 4 and 5 shows that the moss cells are expressing the transmembrane receptor presented in Figure 2 shows a bright green fluorescence as is characteristic for GFP. This fluorescence is mainly located at the plasma membrane which would indicate that our receptor proteins have the correct localization. In comparison to the the transgenic moss expressing GFP in the cytoplasm, the fluorescence of the moss carrying the transmembrane receptor seems to be more associated to the plasma membrane. Hence the fluorescence is a first indication that our receptor works properly but cannot serve as a reliable evidence. Therefore we decided to execute a second method for the proof of localization of our transmembrane receptor.
Verification of receptor orientation by shedding off the ecto-NanoLuc luciferase with TEV protease
We want to investigate the localisation of our synthetic receptor. For this purpose we used a transgenic moss strain which contains NanoLuc luciferase at the ecto-domain of the receptor. Between the luciferase and the transmembrane region, we have added a linker consisting of a Streptag II and a TEV cleavage site. By adding the recombinant TEV protease, the TEV protease should be able to pass the cell wall and enter the appoplast, cleave the TEV cleavage site and cut off the NanoLuc luciferase. Afterwards the luciferase in the supernatant can be analyzed by its luminescense.
To prove this process, we cut off our nLuc-Strep-tag II-construct by using the TEV-Protease. As we hypothetically know, our membrane construct is localized in the membrane and the nLuc-Strep-tag II-construct orientated to the periplasma, protected by the cell-wall of our moss. In conclusion to that, the TEV protease needs to pass the cell-wall to cut off the nLuc-Strep-tag II-construct. This process is possible in liquid medium, because the TEV protease just has a molecular weight between 25 kDa and 27 kDa, so it can pass the cell-wall without hindrance. There, it cuts off the nLuc-Strep-tag II-construct at the TEV cleavage site. Again, this construct has in sum a molecular weight of 20 kDa (Nanoluciferase – 19 kDa, Streptag – 1 kDa), so it can pass the cell-wall due to diffusion to get to the exterior of the moss cell.
After this experimental part, it is necessary to prove the theoretical deliberation. In the following we performed several experiments like SDS-Pages, Luciferase Assays and tried to bind our construct by utilizing different beads.
Experimental Setup
First Step
The reagents which were necessary to perform the first step of the experiment have been 1.863 ml TEV Protease Buffer, 137 µl TEV Protease (0.364 mg/ml) to get a total enzyme activity of 500 Units in each sample and as much genetically modified moss, containing the membrane bound Streptag-Luciferase-Construct, as possible. All reagents have been added to a 2 ml Tube, gently mixed and incubated for 16 hours at 4°C. A negative control containing wild type moss instead of the transgenic one was prepared, too. Before starting the incubation of the prepared samples for 16 hours, we extracted 100 µl of each one, to enable the comparison of the amount of the cut-off construct at the beginning of the incubation time and the incubation over night.
Second Step
In the second step both samples, incubated for 16 hours, were centrifuged at 10000 rpm for 5 minutes to make the moss cells form a pellet at the bottom of the tube. Afterwards the supernatants, where the Nanoluciferase-Streptag-Construct was expected to be found, were transferred to new 2 ml tubes.
Third step
Afterwards we measured the Nanoluciferase activity in the supernatants of the transgenic moss at the beginning of the incubation time and after 16 hours. The same procedure was performed for the Wild-type moss. Additionally we measured the activity of the TEV-Protease buffer in the Luciferase-Assay, to be sure that it does not falsify our results. All samples were prepared with 50 µl supernatant mixed to 50 µl Luciferase-Substrate-Buffer. We concluded that the Luciferase activity was significant in our membrane-construct supernatants, whereas the wild-type moss did not show any significant Luciferase activity.
| Supernatant transgenic moss - T0 | Supernatant transgenic moss - Over night | Supernatant wild-type moss - T0 | Supernatant wild-type moss - Over night | TEV Buffer |
|---|---|---|---|---|
| 137047 | 1852775 | 5 | 20 | 3 |
Conclusion
The Luciferase-Assay showed a significant activity rate, so there has to be a significant amount of cut-off Streptag-Nanoluciferase-Construct in the transgenic supernatant. Due to the significant results of the Luciferase-Assay of the transgenic moss supernatant, the prove of the existence and the desired orientation of our Membrane-Nanoluciferase-Construct is confirmed.
SpyCatcher & SpyTag
As you can clearly see, all the combinations of orientations of SpyTag with SYFP2 lead to a post-translational covalent isopeptide formation to SpyCatcher (Figure 8). Even fusion to both termini of SpyTag leads to a post-translational fusion to SpyCatcher. So with the SpyCatcher/SpyTag system you can not only post-translationally fuse the C-terminus of the first protein to the N-terminus of another protein; also the system allows post-translational non-canonical protein fusions, like C-C terminal, and branched fusions, which is useful for creating two-step enzyme assembly lines.
So by using this system, you can fuse proteins post-translationally in every desired orientation which offers you new oppurtunities in protein engineering.
References
http://www.ncbi.nlm.nih.gov/pubmed/19021876 Gitzinger et al., 2009 Functional cross-kingdom conservation of mammalian and moss (Physcomitrella patens) transcription, translation and secretion machineries, Plant Biotechnol J. 2009 Jan;7(1):73-86
http://www.ncbi.nlm.nih.gov/pubmed/22366317 Zakeri et al., 2012 Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 20;109(12)
 "
"





AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351