Team:Evry/Protocols/01
From 2013.igem.org
| (46 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:Evry/ | + | {{:Team:Evry/template_notebook}} |
<html> | <html> | ||
| Line 5: | Line 5: | ||
<div id="mainTextcontainer"> | <div id="mainTextcontainer"> | ||
| - | |||
| - | <h1 | + | <h1> Competent cells </h1> |
| - | < | + | <h2> Goal </h2> |
| + | <p> | ||
| + | Bacteria can integrate DNA fragments from the environment. <i>Escherichia coli</i>, contrary to <i>Bacillus subtillis</i> another bacteria frequently used in molecular and synthetic biology, is not naturally in a state of competence (able to integrate DNA fragments during <a href='https://2013.igem.org/Team:Evry/Protocols/02' target='_blank'>transformation</a>), that is why we have to prepare them.<br> | ||
| + | Two methods can be used:<br> | ||
| + | -Chimiocompetent cells<br> | ||
| + | -Electrocompetent cells<br> | ||
| - | < | + | We choose to use chimiocompetent cells for usual transformation and electrocompetent cells just for ΔFur strain preparation by <a href="https://2013.igem.org/Team:Evry/Protocols/10" target='_blank'>homologous recombination</a>.<br> |
| + | <h2> Preparation </h2> | ||
| - | < | + | <p>Grow overnight pre-culture of the strain you want to make competent (chimio or electro) in 2 mL of LB.</p> |
| - | + | ||
| - | < | + | <h3>Chimiocompetent cells</h3> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <h4>Solutions preparation</h4> |
| - | + | ||
| - | < | + | <p><b>Solution for 1M Cacl2:</b><br/> |
| - | + | Add 14,30g of CaCl2 into 100 ml distilledwater<br/> | |
| + | </p> | ||
| - | <b> | + | <b><p>Solution for 0,1M Cacl2:</b><br/> |
| - | Add | + | Add 50 mL of CaCl2 1M solution into 450 ml of distilled water<br/> |
| + | </p> | ||
| + | |||
| + | |||
| + | <p><b>Solution for 0,1M Cacl2 + 15% glycerol:</b><br/> | ||
| + | Add 50 mL of CaCl2 1M solution and 75 mL of glycerol 100% into 450 ml of dissilted water<br/> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h4>Competent cells preparation</h4> | ||
| + | <p>For 200 ml LB medium, add 400 µL of strain sample.<br/> | ||
| + | Let the bacteria grow until it reaches an Optical Density (OD) between 0,3 and 0,35. Chimiocompetent bacteria are more able to transform when they are in exponential phase.<br/> | ||
| + | Once it reached the right OD, put the medium on ice for 30 minutes to slow down growth.<br/> | ||
| + | Split the 200 mL into 4x50 mL tubes then centrifuge at 3000 rpm for 5 minutes at 4°C and suppress supernatant afterwards.<br/> | ||
| + | Resuspend the pellet cells with 5 mL of Cacl2 at 0,1M for each 50 mL tube.<br/> | ||
| + | Again, put the medium on ice for 30 minutes.<br/> | ||
| + | Centrifuge at 3000 rpm for 5 minutes at 4°C then suppress supernatant.<br/> | ||
| + | Resuspend the cells with 1 mL of Cacl2 at 0,1M + 15% glycerol for each 50 mL tube.<br/> | ||
| + | Split the total 4 mL into 40 tubes containing each 100 µL of concentrated cell solution. This step should be executed fast enough and on ice.<br/> | ||
| + | Store at - 80°C.</p> | ||
| + | |||
| + | |||
| + | <h3>Electrocompetent cells</h3> | ||
| + | <p>For 200 ml LB medium, add 200 µL of strain sample.<br/> | ||
| + | Let the bacteria grow until it reaches an OD between 0,5 and 0,7.<br/> | ||
| + | Once it reached the right OD, put the medium on ice for 20 minutes to slow down growth.<br/> | ||
| + | Split the 200 mL into 4x50 mL tubes then centrifuge at 4000 rpm for 15 minutes at 4°C and suppress supernatant afterwards.<br/> | ||
| + | Resuspend the pellet cells with 50 mL of ice-cold 10% glycerol for each 50 mL tube.<br/> | ||
| + | Centrifuge at 4000 rpm for 15 minutes at 4°C then suppress supernatant.<br/> | ||
| + | Resuspend the cells with 25 mL of ice-cold 10% glycerol for each 50 mL tube.<br/> | ||
| + | Centrifuge at 4000 rpm for 15 minutes at 4°C then suppress supernatant.<br/> | ||
| + | Resuspend the cells with 2 mL of ice-cold 10% glycerol for each 50 mL tube.<br/> | ||
| + | Centrifuge at 4000 rpm for 15 minutes at 4°C then suppress supernatant.<br/> | ||
| + | Resuspend the cells with 2 mL of ice-cold 10% glycerol for each 50 mL tube.<br/> | ||
| + | Split the total 4 mL into 40 tubes containing each 100 µL of concentrated cell solution. This step should be executed fast enough and on ice.<br/> | ||
| + | Store at - 80°C.</p> | ||
| + | |||
| + | |||
| + | <h2>Contamination and competence tests</h2> | ||
| + | |||
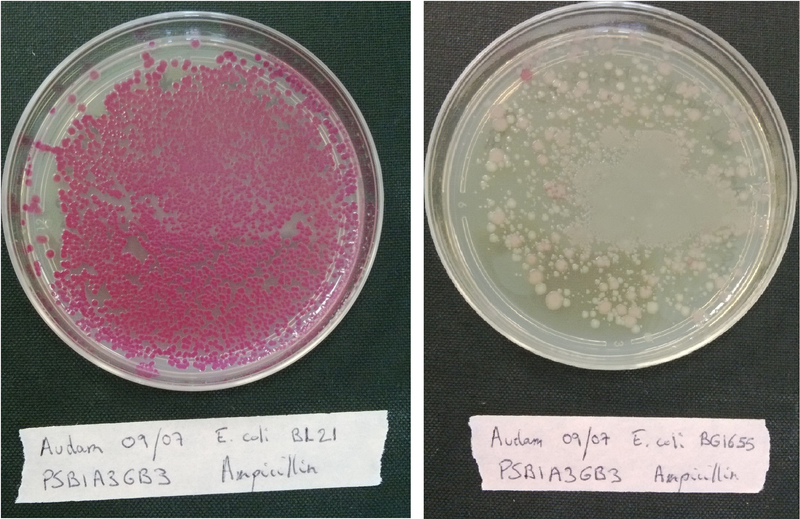
| + | <p>Plate each strain on LB medium with Ampicillin, Kanamycin or Chloramphenicol in order to evaluate if it contaminated or not. <br/> | ||
| + | Transform each strain with a pSB1A3 plasmid (red colonies) and plated them on LB medium with Ampicillin only to evaluate wether our strains are competent or not.<br> | ||
| + | If the bacteria is competent, it will incorporate the plasmid that code for a red protein and thus have a red phenotype.(see figure below)</p> | ||
| + | |||
| + | |||
| + | |||
| + | <div align="center"> | ||
| + | <div class="captionedPicture" style="width:40%;float:center;"> | ||
| + | <a title="Competence test" href="https://static.igem.org/mediawiki/2013/thumb/9/99/Competence_petri_dish.png/800px-Competence_petri_dish.png"> | ||
| + | <img alt="Competence test" src="https://static.igem.org/mediawiki/2013/thumb/9/99/Competence_petri_dish.png/800px-Competence_petri_dish.png" class="Picture"/> | ||
| + | </a> | ||
| + | <div class="caption"> | ||
| + | <b>Figure 1:</b> <i>Escherichia coli</i> (MG1655 strain) on the left are highly competent and expressed RFP, bacteria on the right are little competent. | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
Latest revision as of 13:26, 1 October 2013
Competent cells
Goal
Bacteria can integrate DNA fragments from the environment. Escherichia coli, contrary to Bacillus subtillis another bacteria frequently used in molecular and synthetic biology, is not naturally in a state of competence (able to integrate DNA fragments during transformation), that is why we have to prepare them.
Two methods can be used:
-Chimiocompetent cells
-Electrocompetent cells
We choose to use chimiocompetent cells for usual transformation and electrocompetent cells just for ΔFur strain preparation by homologous recombination.
Preparation
Grow overnight pre-culture of the strain you want to make competent (chimio or electro) in 2 mL of LB.
Chimiocompetent cells
Solutions preparation
Solution for 1M Cacl2:
Add 14,30g of CaCl2 into 100 ml distilledwater
Solution for 0,1M Cacl2:
Add 50 mL of CaCl2 1M solution into 450 ml of distilled water
Solution for 0,1M Cacl2 + 15% glycerol:
Add 50 mL of CaCl2 1M solution and 75 mL of glycerol 100% into 450 ml of dissilted water
Competent cells preparation
For 200 ml LB medium, add 400 µL of strain sample.
Let the bacteria grow until it reaches an Optical Density (OD) between 0,3 and 0,35. Chimiocompetent bacteria are more able to transform when they are in exponential phase.
Once it reached the right OD, put the medium on ice for 30 minutes to slow down growth.
Split the 200 mL into 4x50 mL tubes then centrifuge at 3000 rpm for 5 minutes at 4°C and suppress supernatant afterwards.
Resuspend the pellet cells with 5 mL of Cacl2 at 0,1M for each 50 mL tube.
Again, put the medium on ice for 30 minutes.
Centrifuge at 3000 rpm for 5 minutes at 4°C then suppress supernatant.
Resuspend the cells with 1 mL of Cacl2 at 0,1M + 15% glycerol for each 50 mL tube.
Split the total 4 mL into 40 tubes containing each 100 µL of concentrated cell solution. This step should be executed fast enough and on ice.
Store at - 80°C.
Electrocompetent cells
For 200 ml LB medium, add 200 µL of strain sample.
Let the bacteria grow until it reaches an OD between 0,5 and 0,7.
Once it reached the right OD, put the medium on ice for 20 minutes to slow down growth.
Split the 200 mL into 4x50 mL tubes then centrifuge at 4000 rpm for 15 minutes at 4°C and suppress supernatant afterwards.
Resuspend the pellet cells with 50 mL of ice-cold 10% glycerol for each 50 mL tube.
Centrifuge at 4000 rpm for 15 minutes at 4°C then suppress supernatant.
Resuspend the cells with 25 mL of ice-cold 10% glycerol for each 50 mL tube.
Centrifuge at 4000 rpm for 15 minutes at 4°C then suppress supernatant.
Resuspend the cells with 2 mL of ice-cold 10% glycerol for each 50 mL tube.
Centrifuge at 4000 rpm for 15 minutes at 4°C then suppress supernatant.
Resuspend the cells with 2 mL of ice-cold 10% glycerol for each 50 mL tube.
Split the total 4 mL into 40 tubes containing each 100 µL of concentrated cell solution. This step should be executed fast enough and on ice.
Store at - 80°C.
Contamination and competence tests
Plate each strain on LB medium with Ampicillin, Kanamycin or Chloramphenicol in order to evaluate if it contaminated or not.
Transform each strain with a pSB1A3 plasmid (red colonies) and plated them on LB medium with Ampicillin only to evaluate wether our strains are competent or not.
If the bacteria is competent, it will incorporate the plasmid that code for a red protein and thus have a red phenotype.(see figure below)
 "
"