Team:Bielefeld-Germany/Biosafety/Biosafety System S
From 2013.igem.org
| Line 1: | Line 1: | ||
{{Team:Bielefeld-Germany/Header2}} | {{Team:Bielefeld-Germany/Header2}} | ||
{{Team:Bielefeld-Germany/css/header_cleanup.css}} | {{Team:Bielefeld-Germany/css/header_cleanup.css}} | ||
| + | {{Team:Bielefeld-Germany/css/button.css}} | ||
| + | |||
| + | |||
__NOTOC__ | __NOTOC__ | ||
<html> | <html> | ||
| - | + | <style> | |
| - | + | h1{padding:10px 0;padding-left:192px; padding-right:192px; margin-top:70px; } | |
| - | + | .toc, #toc{width:255px;font-size:85%;} | |
| + | |||
| + | #globalwrapper ul {padding-left:40px; padding-right:40px;} | ||
| + | #globalwrapper #rightcol ul {padding-left:0px; padding-right:0px;} | ||
| + | |||
| + | h2,h3,h4{clear:both;} | ||
| + | #globalwrapper h4{color:#ff6600; padding-left:20px;} | ||
| + | #globalwrapper div.thumb.tleft{margin-left:20px; margin-right:20px; clear:both;} | ||
| + | |||
| + | #globalwrapper ul {clear:both;} | ||
| + | #globalwrapper ul ul{clear:both;} | ||
| + | #globalwrapper ul{padding-left:60px; padding-right:40px;} | ||
| + | #globalwrapper ul ul{padding-left:40px; padding-right:40px;} | ||
| + | |||
| + | #globalwrapper p{padding-left:0px; padding-right:40px;} | ||
| + | #globalwrapper .bigbutton p{padding-left:5px; padding-right:5px; padding-top:2px;} | ||
| + | |||
| + | .bigbutton{width:150px; height:50px; line-height:50px; font-size:1.2em; margin-right:10px; display:table;} | ||
| + | .bigbutton a{display:block; height:100%;} | ||
| + | |||
| + | .rightcol{ | ||
| + | width:140px; | ||
| + | height:450px; | ||
| + | margin-left:800px; | ||
| + | float:right; | ||
| + | position:fixed; | ||
| + | margin-top:0px; | ||
| + | overflow-y:scroll; | ||
| + | box-shadow:0px 0px 2px 0px grey; | ||
| + | padding:0px 20px; | ||
| + | |||
| + | </style> | ||
</html> | </html> | ||
| - | < | + | <div id=globalwrapper style="padding-left:20px; padding-right:20px"> |
| - | + | <div id="leftcol" style="width:750px; float:left; overflow:auto;"> | |
| - | < | + | <html> |
| + | <h1>Biosafety System AraCtive</h1> | ||
| + | <div id="buttonrow" style="padding-top:30px; padding-bottom:70px; padding-left:45px; clear:both;"> | ||
| - | + | <div class="bigbutton"> | |
| + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Biosafety">Biosafety Overview</a></div> | ||
| + | <div class="bigbutton"> | ||
| + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System">System Overview</a></div> | ||
| + | <div class="bigbutton"> | ||
| + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S#Genetic_Approach">Genetic Approach</a></div> | ||
| + | <div class="bigbutton"> | ||
| + | <a href="https://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S#Results">Results</a></div> | ||
| - | < | + | </div> |
| - | + | </html> | |
| - | + | ||
| - | + | ||
| + | ==Overview== | ||
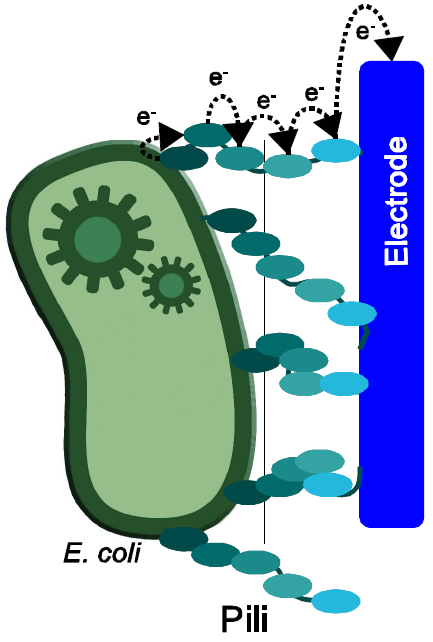
| - | [[ | + | [[Image:Bielefeld-germany-project-overview-nanowires.png|left|thumb|250px|'''Figure 1:''' Principle of electron transfer from bacteria to anode via nanowires.]] |
| - | < | + | <p align="justify"> |
| + | The tetracyclin repressor (TetR)/ operator (TetO) originally is used by E. coli to work against the antibiotic tetracycline but in many cases it is used for regulated expression for industrial processes. When there is no tetracycline available the TetR binds with high affinity the tetracycline operator. When tetracycline is available the TetR switches his conformation and so it comes to a dissolution of the TetR and the TetO. Because of this the polymerase isn’t enhanced anymore and is able to express the genes which lies behind the TetO. In our system the TetR is under the control of a rhamnose promotor (rha-promotor) which only works in the presence of rhamnose. When the bacteria would break out of the MFC there wouldn’t be enough rhamnose in the environment to activate the promotor in a way that enough TetR would be produced to block the polymerase by binding at the TetO. Therefore the polymerase binds to the promotor of TetO and it comes to the expression of RNase Ba and the degradation of the DNA. | ||
| + | </p> | ||
| - | |||
| - | |||
| - | |||
<br><br> | <br><br> | ||
| - | |||
| - | |||
| - | + | ||
| + | ==Genetic Approach== | ||
| + | |||
[[File:IGEM Bielefeld 2013 Biosafety System S.png|600px|thumb|center|System S in the MFC: In this case the mikroorganism is in the MFC with sufficient L-rhamnose. It comes to an expression of araC which blocks the arabinose-promoter by binding and alr which switches L-alanine to D-alanine. Because of the fact that araC blocks the arabinose-promoter the RNase Ba can't expressed.]] | [[File:IGEM Bielefeld 2013 Biosafety System S.png|600px|thumb|center|System S in the MFC: In this case the mikroorganism is in the MFC with sufficient L-rhamnose. It comes to an expression of araC which blocks the arabinose-promoter by binding and alr which switches L-alanine to D-alanine. Because of the fact that araC blocks the arabinose-promoter the RNase Ba can't expressed.]] | ||
| Line 47: | Line 88: | ||
[[File:IGEM Bielefeld 2013 Biosafety System S ohne Rhamnose.png|600px|thumb|center|System S outside of the MFC: In this case the mikroorganism could get out of the MFC by damage or incorrect handling. Outside of the MFC there isn't enough L-rhamnose. So araC doesn't block the arabinose-promotor any more and RNase Ba can be expressed.E.coli dies.]] | [[File:IGEM Bielefeld 2013 Biosafety System S ohne Rhamnose.png|600px|thumb|center|System S outside of the MFC: In this case the mikroorganism could get out of the MFC by damage or incorrect handling. Outside of the MFC there isn't enough L-rhamnose. So araC doesn't block the arabinose-promotor any more and RNase Ba can be expressed.E.coli dies.]] | ||
| + | |||
| + | |||
| + | [[File:IGEM Bielefeld 2013 biosafety Rhamnose-promoter.png]] | ||
| + | |||
| + | [[File:IGEM Bielefeld 2013 biosafety araC.png]] | ||
| + | |||
| + | [[File:IGEM Bielefeld 2013 biosafety alr.png]] | ||
| + | |||
| + | [[File:IGEM Bielefeld 2013 biosafety Terminator.png]] | ||
| + | |||
| + | [[File:IGEM Bielefeld 2013 biosafety pBAD.png]] | ||
| + | |||
| + | [[File:IGEM Bielefeld 2013 biosafety RNase Ba.png]] | ||
| + | |||
| + | |||
| + | ==Results== | ||
| + | |||
| + | |||
| + | |||
| + | ==References== | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br><br><br><br> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div id="rightcol" style="width:210px; height:450px; margin-left:752px; margin-top:33px; float:right; position:fixed; overflow-y:scroll; box-shadow:0px 0px 2px 0px grey;" padding:0px 20px;> | ||
| + | __TOC__ | ||
| + | <div id="spacer" style="height:300px"></div> | ||
| + | </div> | ||
</div> | </div> | ||
Revision as of 23:04, 29 September 2013
Biosafety System AraCtive
Overview
The tetracyclin repressor (TetR)/ operator (TetO) originally is used by E. coli to work against the antibiotic tetracycline but in many cases it is used for regulated expression for industrial processes. When there is no tetracycline available the TetR binds with high affinity the tetracycline operator. When tetracycline is available the TetR switches his conformation and so it comes to a dissolution of the TetR and the TetO. Because of this the polymerase isn’t enhanced anymore and is able to express the genes which lies behind the TetO. In our system the TetR is under the control of a rhamnose promotor (rha-promotor) which only works in the presence of rhamnose. When the bacteria would break out of the MFC there wouldn’t be enough rhamnose in the environment to activate the promotor in a way that enough TetR would be produced to block the polymerase by binding at the TetO. Therefore the polymerase binds to the promotor of TetO and it comes to the expression of RNase Ba and the degradation of the DNA.
Genetic Approach

Results
References
 "
"