Team:Bielefeld-Germany/Project/Cytochromes
From 2013.igem.org
| Line 15: | Line 15: | ||
<font face=""> | <font face=""> | ||
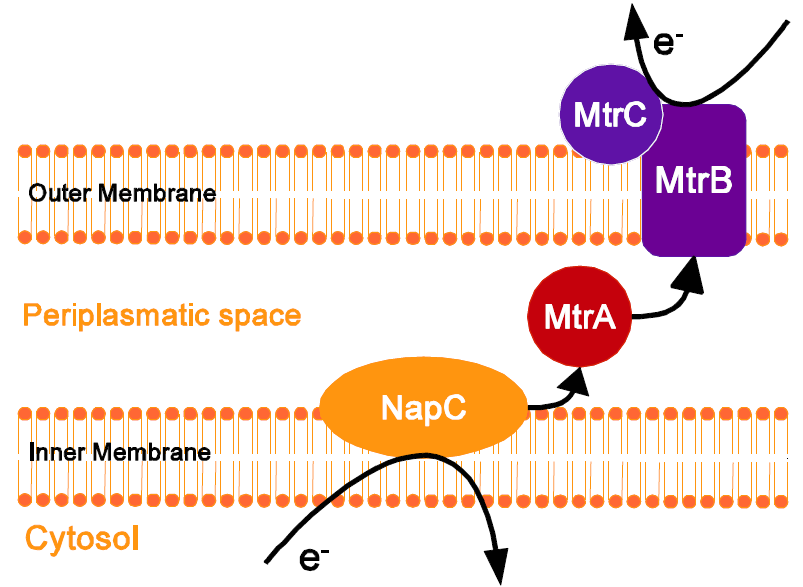
| - | + | Cell membranes work as a natural insulator and prevent the flow from electrons out of the cell. To enable transfer of electrons from the general metabolism to the outside of the cell we had to alter the membrane of our organism E. coli without disturbing cell growth, stability and metabolism. Some species from the genera Shewanella and Geobacter have developed different mechanisms to allow extracellular electron transfer. In Shewanella oneidensis MR-1 this is achieved via different c-type cytochromes, which shuttle the electrons along a defined molecular route from the cytoplasma and the inner membrane to the outside of the cell during anaerobic respiration. This pathway is very well understood and characterized []. | |
| - | + | <br> | |
| + | Previous work suggests that a working electron transfer chain can be achieved by a minimal set of three genes, tthe periplasmatic decaheme MtrA, the outer membrane β -barrel protein mtrB and the outer membrane cytochromes MtrC. MtrA interacts with at least one native redox protein, f.e. CymA and can therefore start the transfer of electrons. | ||
| + | <br> | ||
| + | Additionally another set of genes, the cytochrome c maturation genes (ccmABCDEFGH), is required for correct protein localization and heme insertion into MtrA and MtrC. Under anaerobic conditions these genes are naturally expressed in E. coli, whereas under aerobic conditions we had to co-express them. For aerobic growth the cells were transformed with both plasmids, containing the cytochrome-cluster, as well as the ccm-cluster . By this approach extracellular electron transfer should be possible in E.coli and allow the use of this genetically engineered strain in a microbial fuel cell. | ||
| + | |||
Revision as of 21:21, 18 September 2013
Mediators
Cell membranes work as a natural insulator and prevent the flow from electrons out of the cell. To enable transfer of electrons from the general metabolism to the outside of the cell we had to alter the membrane of our organism E. coli without disturbing cell growth, stability and metabolism. Some species from the genera Shewanella and Geobacter have developed different mechanisms to allow extracellular electron transfer. In Shewanella oneidensis MR-1 this is achieved via different c-type cytochromes, which shuttle the electrons along a defined molecular route from the cytoplasma and the inner membrane to the outside of the cell during anaerobic respiration. This pathway is very well understood and characterized [].
Previous work suggests that a working electron transfer chain can be achieved by a minimal set of three genes, tthe periplasmatic decaheme MtrA, the outer membrane β -barrel protein mtrB and the outer membrane cytochromes MtrC. MtrA interacts with at least one native redox protein, f.e. CymA and can therefore start the transfer of electrons.
Additionally another set of genes, the cytochrome c maturation genes (ccmABCDEFGH), is required for correct protein localization and heme insertion into MtrA and MtrC. Under anaerobic conditions these genes are naturally expressed in E. coli, whereas under aerobic conditions we had to co-express them. For aerobic growth the cells were transformed with both plasmids, containing the cytochrome-cluster, as well as the ccm-cluster . By this approach extracellular electron transfer should be possible in E.coli and allow the use of this genetically engineered strain in a microbial fuel cell.
 "
"