Shuttle vectors
Building bridges between E. coli and Lactobacillus
A shuttle vector is a plasmid that can be transferred between two different species and is able to replicate in both. Such a vector is key for bioengineering in Lactobacillus. The low transformation frequencies of ligations and longer generation times slows down the work pace tremendously if you have to assembly and test everything in Lactobacillus. A solution to this problem is if you instead build everything in E.coli and then transfer the finished construct to Lactobacillus. We have created two shuttle vectors that we have sent to the registry.
Resveratrol belongs to a group of molecules known as Phyloalexin which is used by many plants to battle infections of all sorts.
Since the early 90’s there has been many studies done on resveratrol showing it has a wide range of beneficial properties ranging from skin cancer reduction to anti-inflamatory effects and antioxidant properties.
Shuttle vectors
BBa_K1033206, with chloramphenicol resistance.
BBa_K1033207, with erythromycin resistance.
Construction of shuttle vectorns
To create the shuttle vector we replaced the replicon in a biobrick compatible plasmid BBa_K864003?) with a replicon from a plasmid known to work in both E.coli and Lactobacillus pSH71. pSH71 originates from a plasmid, pJP059, from Lactococcus lactis but it is well known to replicate in both the Lactobacillus genus and in E.coli through rolling circle replication.[1]
Many species in the Lactobacillus genus have inherent antibiotic resistances (referens nr 2), because of that we have been limited in our selection of antibiotic resistance. Most commonly used are chloramphenicol and erythromycin and we made versions of both for our shuttle vector. BBa_K1033207 contains an erythromycin cassette from pLUL631 (länk till artikel) and BBa_K1033206 contains the resistance cassette from ??? but has had its promoter replaced with our cp29 promoter.

Results
We have managed to transform both shuttle vectors to E. coli and verified them by sequencing. We have also managed to subclone with it and replace the red insert with chromoproteins expressed by our CP promoters that works both in E. coli and Lactobacillus, clearly yielding coloured colonies (see picture).
Biobrick
We succeded in the cloning and sequencing of our two biobricks, 4-Coumarate ligase from
arabidophsis thaliana and Stilbene synthase from vitis vinifiera with the RBS B0034 that should work in various organisms, lactobacillus and e-coli. Sequencing was done at GATC biotech and Uppsala Genome center using sanger sequencing.
We also succeded in the making of our operon containing 4Cl and STS together.
BBa_K1033000 - tyrosine ammonia lyase (TAL) with RBS
BBa_K1033001 - 4 coumarate ligase (4CL) with RBS
BBa_K1033002 - stilbene synthase (STS) with RBS
BBa_K1033003 - B0034-4Cl-B0034-STS
Western blot
We also succeeded in expressing the enzyme stilbene synthase in E. coli. Although our expression of the protein was very weak, and due to time constraints we were not able to optimize our experiment.
To enable the detection of this protein by anti-his antibodies, 6-histidine tags was incorporated in the sequence. This way we could detect our enzyme with anti-his antibodies.
We expressed our protein with a promotor working in both lactobacillus and e-coli. This way, we can
easily transfer stilbene synthase to lactobacillus later on.
The size of our protein was calculated using ProtParam
[5], 43 kDA.
 Figure 1:Number 2 shows a very weak band of our protein at around 43 kDA.
Figure 1:Number 2 shows a very weak band of our protein at around 43 kDA.
Positive control -> 1, Stilbene synthase -> 2
High pressure liquid chromatography
We tested our biobrick 4Cl-STS on HPLC, by adding p-coumaric acid as a precursor. The result we saw was quite unclear. We saw that the e-coli produced something out of the ordinary, but the absorbation was low and the peaks did not exactly match the standard. The peak at around ~33 min could correspond to our standard, but it is unclear. We theorize it is something in the actual hplc measurement that fails, or that something happens to our resveratrol metabolite in our e-coli. This result could correspond to the poor results in our blot. We hope that iGEM teams can continue to work on these biobricks in the future.
4Cl-STS translational fusion expressed in e-coli
 Figure 1: E. coli supposed to produce resveratrol. As we can see, we got very low absorbance peaks at ~30 min, ~33 min and ~36 min.
Resveratrol standard
Figure 1: E. coli supposed to produce resveratrol. As we can see, we got very low absorbance peaks at ~30 min, ~33 min and ~36 min.
Resveratrol standard
 Figure 2: Resveratrol standard, peaks around ~33, ~34 min.
Resveratrol standard scaled
Figure 2: Resveratrol standard, peaks around ~33, ~34 min.
Resveratrol standard scaled
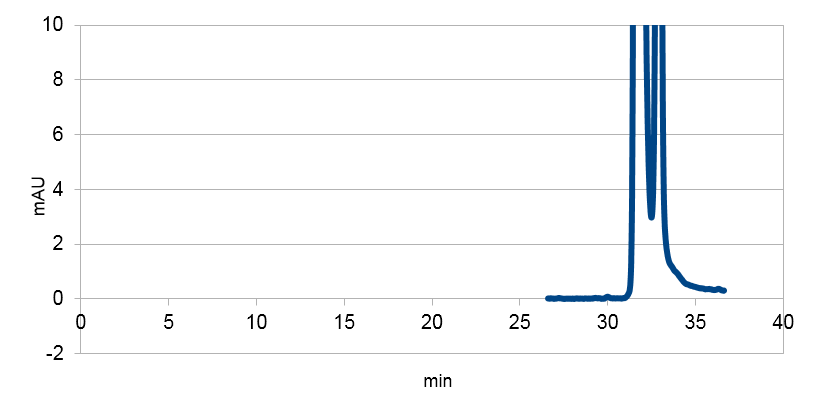 Figure 3: Resveratrol standard that is scaled down to correspond to the absorbations of our e. coli supposed to produce the corresponding metabolite. The peaks are at around ~33 and ~34.
Lysed bacterial culture without plasmid of assembly
Figure 3: Resveratrol standard that is scaled down to correspond to the absorbations of our e. coli supposed to produce the corresponding metabolite. The peaks are at around ~33 and ~34.
Lysed bacterial culture without plasmid of assembly
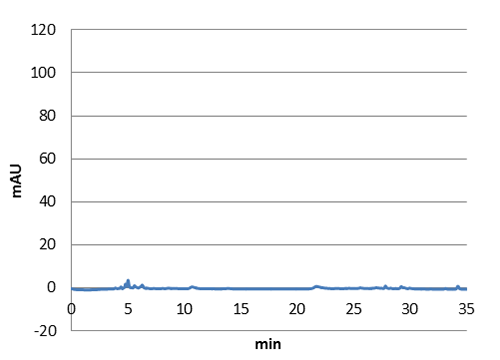 Figure 3: E. coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peaks around 30-35 minutes.
Figure 3: E. coli culture injected to the hplc without our biobrick tyrosine ammonia lyase. Here we can see that there is originally no peaks around 30-35 minutes.
[1] Sinclair, D. A & Baur, Y, A. Therapeutic potential of resveratrol:
the in vivo evidence. Nature 506 | JUNE 2006 | VOLUME 5
[2] Baur, Y, A. et al. Resveratrol improves health and survival
of mice on a high-calorie diet. Nature Vol 444| 16 November 2006
[3] Robert J. Conrado et al, DNA guided assembly of biosynthetic pathways promotes improved catalytic effiency. Nucleic Acids Research , 2012, Vol 40 NO 4, 1879-1889
[4] Sinclair, D. A & Baur, Y, A. Therapeutic potential of resveratrol:
the in vivo evidence. Nature 506 | JUNE 2006 | VOLUME 5
[5] Baur, Y, A. et al. Resveratrol improves health and survival
of mice on a high-calorie diet. Nature Vol 444| 16 November 2006
[6] Here's a link to ProtParam: http://web.expasy.org/protparam/
 "
"















