Team:Bielefeld-Germany/Labjournal/May
From 2013.igem.org
(Difference between revisions)
| Line 5: | Line 5: | ||
__NOTOC__ | __NOTOC__ | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div id=globalwrapper style="padding-left:20px; padding-right:20px"> | <div id=globalwrapper style="padding-left:20px; padding-right:20px"> | ||
| Line 117: | Line 103: | ||
| - | <div id="asdf" | + | <div id="asdf" class="withMenu;"> |
<html> | <html> | ||
<div id="nav2" style="width:210px; padding-bottom:5px; padding-left:15px;"> | <div id="nav2" style="width:210px; padding-bottom:5px; padding-left:15px;"> | ||
| Line 145: | Line 131: | ||
| - | <div id="rightcol" style="width:210px; height: | + | <div id="rightcol" style="width:210px; height:100%; overflow-y:auto; box-shadow:0px 0px 2px 0px grey;" padding:0px 20px;> |
__TOC__ | __TOC__ | ||
<div id="spacer" style="height:300px"></div> | <div id="spacer" style="height:300px"></div> | ||
Revision as of 21:12, 4 October 2013
May
Milestones
- Initiating labwork on the sub-project endogenous mediator Glycerol dehydrogenase.
- Starting lab work with the first successful PCR: GldA with Pre- and Suffix overlaps could be amplified from Escherichia coli genome.
- The main work however is still planning of our subprpjects. Designing experiments and a lot of research.
- Finding sponsors goes ahead. Many companies like our project and want to support us.
Week 1
Organization
- We were already able to find sponsors for our team: IIT BIEKUBA, IIT Biotech, Evonik, PlasmidFactory and FisherScientific will support us.
MFC
Mediators
- Glycerol dehydrogenase
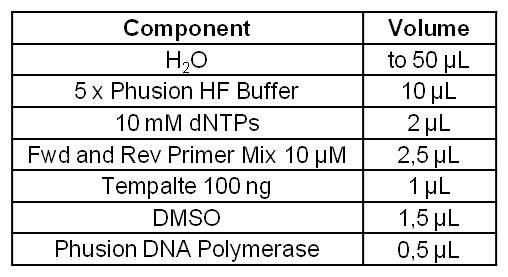
- Primerdesign for isolation of GldA gene from Escherichia coli KRX strain, with overlaps for Biobrick Prefix and Suffix:
- Forward Primer GldA (43 bp): GAATTCGCGGCCGCTTCTAGATGGACCGCATTATTCAATCACC
- Reverse Primer GldA (45 bp): CTGCAGCGGCCGCTACTAGTATTATTCCCACTCTTGCAGGAAACG
2.Week
Organization
- Distribution of our team in various subgroups for best work efficiency.
- We’ve organized a working group barbecue to thank the AG of Dr. Jörn Kalinowski in the CeBiTec for the appropriation of space and support in the laboratory.
MFC
3.Week
Organization
- Planning the trip to the European jamboree in Lyon from 11.-13. October 2013.
MFC
Mediators
- Glycerol dehydrogenase
- Starting first cultivation of Escherichia coli KRX strain for complete genome isolation.
- Successful genome isolation of Escherichia coli KRX.
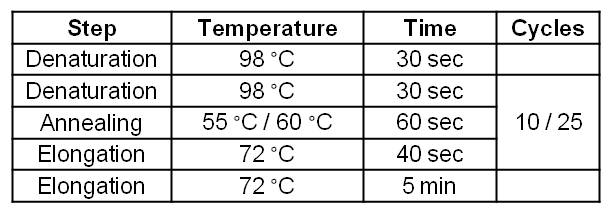
- Successful PCR on the GldA gene of Escherichia coli KRX strain.
- GldA PCR product was isolated by Agarose gel electrophorese and purificated.
- Bands are at expected size of 1050 bp.

Figure 1: Agarosegel from PCR on the GldA gene of Escherichia coli KRX strain with forward and reverse primer GldA. For Ladder we used [http://www.thermoscientificbio.com/nucleic-acid-electrophoresis/generuler-1-kb-dna-ladder-ready-to-use-250-to-10000-bp GeneRuler™ 1 kb DNA Ladder fromThermo Scientific].
4.Week
Organization
- We will also take part at the congress "Next generation of biotechnological processes 2020+" in Berlin at 27. June 2013.
- Having a second presentation at Merck KGaA head office in Darmstadt for explaining our project in detail. We are happy that Merck will be our next supporter in 2013.
MFC
 "
"