Team:Bielefeld-Germany/Labjournal/June
From 2013.igem.org
(Difference between revisions)
| Line 16: | Line 16: | ||
| - | *Starting | + | *Starting lab work on the sub-project [[Team:Bielefeld-Germany/Project/Porins|Porins]]. |
*Successful PCR on OprF gene from'' Pseudomonas fluorescens''. OprF with Pre- and Suffix overlaps could be amplified from genome. | *Successful PCR on OprF gene from'' Pseudomonas fluorescens''. OprF with Pre- and Suffix overlaps could be amplified from genome. | ||
*Planning of our [[Team:Bielefeld-Germany/HumanPractice|Human Practice]] projects started and the first participations are fixed. | *Planning of our [[Team:Bielefeld-Germany/HumanPractice|Human Practice]] projects started and the first participations are fixed. | ||
| Line 36: | Line 36: | ||
====Porines==== | ====Porines==== | ||
| - | * | + | *Primer design for isolation of OprF from ''Pseudomonas fluorescens'' strain, with overlaps for BioBrick Prefix and Suffix: |
*Forward Primer OprF (49 bp): GAATTCGCGGCCGCTTCTAGATGAAACTGAAAAACACCTTGGGCTTTGC | *Forward Primer OprF (49 bp): GAATTCGCGGCCGCTTCTAGATGAAACTGAAAAACACCTTGGGCTTTGC | ||
*Reverse Primer OprF (51 bp): CTGCAGCGGCCGCTACTAGTATTACTTAGCTTGGGCTTCAACCTGCGCTTC | *Reverse Primer OprF (51 bp): CTGCAGCGGCCGCTACTAGTATTACTTAGCTTGGGCTTCAACCTGCGCTTC | ||
| Line 42: | Line 42: | ||
====Nanowires==== | ====Nanowires==== | ||
| - | * | + | *Primer design for isolation of gene-cluster GSU 1491-1495, GSU 1496-1505 and GSU Promoter-1496-1505 from ''Geobacter sulfurreducens'', containing overlaps for BioBrick Prefix and Suffix: |
**Forward GSU 1491-1495 (45 bp):<br> GAATTCGCGGCCGCTTCTAGATGCAGGCTAGCAGACTGGGAGAAC | **Forward GSU 1491-1495 (45 bp):<br> GAATTCGCGGCCGCTTCTAGATGCAGGCTAGCAGACTGGGAGAAC | ||
**Reverse GSU 1491-1495 (38 bp): <br>CTGCAGCGGCCGCTACTAGTATCACTCCTCATCCATGC | **Reverse GSU 1491-1495 (38 bp): <br>CTGCAGCGGCCGCTACTAGTATCACTCCTCATCCATGC | ||
| Line 54: | Line 54: | ||
====Organization==== | ====Organization==== | ||
| - | *Thanks to NEB | + | *Thanks to NEB for the [http://www.neb-online.de/index.php/en/neb-announcements/231-igem-2013 free iGEM kit] with many useful laboratory things for all german iGEM teams. |
*We are working on our first press release. | *We are working on our first press release. | ||
*Having a short radio contribution in the Bielefeld university campus radio ([http://www.radiohertz.de/beta-site radio 87.9 hertz]). | *Having a short radio contribution in the Bielefeld university campus radio ([http://www.radiohertz.de/beta-site radio 87.9 hertz]). | ||
| Line 82: | Line 82: | ||
| - | *Cultivation of Shewanella oneidensis MR-1 in liquid LB medium at 30 °C | + | *Cultivation of ''Shewanella oneidensis'' MR-1 in liquid LB medium at 30 °C |
| - | *Isolation of genomic DNA from S. oneidensis and dilution to the subsequently used PCR template: | + | *Isolation of genomic DNA from ''S. oneidensis'' and dilution to the subsequently used PCR template: |
**4-2006-453: 5.5ng/µl | **4-2006-453: 5.5ng/µl | ||
| Line 90: | Line 90: | ||
*Amplification of the mtrCAB cluster with [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes Phusion polymerase] | *Amplification of the mtrCAB cluster with [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/Molecular#Used_enzymes Phusion polymerase] | ||
**Annealing: Gradient [55.8 - 56.7 - 57.8 - 59.1 - 60.4 - 61.7 - 62.9 - 63.9] | **Annealing: Gradient [55.8 - 56.7 - 57.8 - 59.1 - 60.4 - 61.7 - 62.9 - 63.9] | ||
| - | **Elongation: 1: | + | **Elongation: 1:15 min |
| - | **Notes: Clear Bands at the expected 5. | + | **Notes: Clear Bands at the expected 5.2 kb, the annealing temperature seems not to have an effect. |
| Line 106: | Line 106: | ||
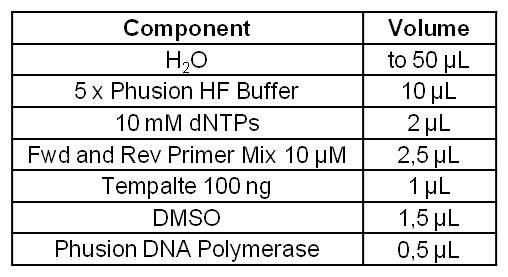
[[Image:IGEM_Bielefeld_Standard_Phusion_PCR_Master_MixLRO.jpg|300px|thumb|left|<p align="justify"> '''Table 1: Standard Phusion PCR Master Mix. '''</p>]] | [[Image:IGEM_Bielefeld_Standard_Phusion_PCR_Master_MixLRO.jpg|300px|thumb|left|<p align="justify"> '''Table 1: Standard Phusion PCR Master Mix. '''</p>]] | ||
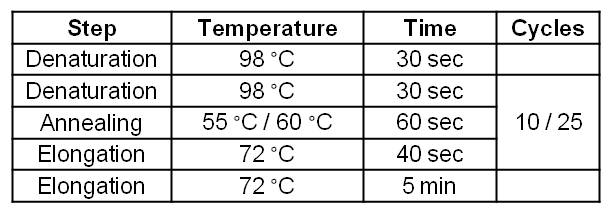
| - | [[Image:IGEM_Bielefeld_Standard_Phu_PCR_GldA_OprF.jpg|300px|thumb|center| <p align="justify">'''Table 2: Two step standard Phusion PCR | + | [[Image:IGEM_Bielefeld_Standard_Phu_PCR_GldA_OprF.jpg|300px|thumb|center| <p align="justify">'''Table 2: Two step standard Phusion PCR program for GldA amplification. '''</p>]] |
| - | *OprF PCR product was isolated by | + | *OprF PCR product was isolated by agarose gel electrophoresis and [[Team:Bielefeld-Germany/Labjournal/Molecular#Used Kits | purified]]. |
*Bands are at expected size of 1300 bp. | *Bands are at expected size of 1300 bp. | ||
| - | [[Image:IGEM_Bielefeld_OprF_standard_PCR.jpg|200px|thumb|left| <p align="justify">'''Figure 1: | + | [[Image:IGEM_Bielefeld_OprF_standard_PCR.jpg|200px|thumb|left| <p align="justify">'''Figure 1: Agarose gel from PCR on the OprF gene of ''Pseudomonas fluorescens'' strain with forward and reverse primer OprF. For Ladder we used [http://www.thermoscientificbio.com/nucleic-acid-electrophoresis/generuler-1-kb-dna-ladder-ready-to-use-250-to-10000-bp GeneRuler™ 1 kb DNA Ladder from Thermo Scientific]. '''</p>]] |
<br> | <br> | ||
| Line 119: | Line 119: | ||
====Nanowires==== | ====Nanowires==== | ||
| - | *Anaerobic cultivation of ''Geobacter sulfurreducens'' strain DSM-12127 in nitrogen-gassed [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/Molecular#Geobacter_Medium ''Geobacter''-medium] , which was suggested by the strain-supplier: German Collection of Microorganisms and Cell Cultures DSMZ, using 30 mL cultivation-tubes and silicone stoppers with upending rim. | + | *Anaerobic cultivation of ''Geobacter sulfurreducens'' strain DSM-12127 in nitrogen-gassed [https://2013.igem.org/Team:Bielefeld-Germany/Labjournal/Molecular#Geobacter_Medium ''Geobacter''-medium], which was suggested by the strain-supplier: German Collection of Microorganisms and Cell Cultures DSMZ, using 30 mL cultivation-tubes and silicone stoppers with upending rim. |
<br><br><br><br> | <br><br><br><br> | ||
<br><br><br><br> | <br><br><br><br> | ||
| Line 131: | Line 131: | ||
====Organization==== | ====Organization==== | ||
| - | *Let’s go to Lyon, flights are booked for the [https://2013.igem.org/Europe/Pre-Jamboree European jamboree in Lyon] from 11 | + | *Let’s go to Lyon, flights are booked for the [https://2013.igem.org/Europe/Pre-Jamboree European jamboree in Lyon] from October 11 to 13, 2013. |
| - | *We will participate at ‘[http://www.bio.nrw.de/studentconvention BioNRW pHD Student Convention]’ in Düsseldorf at 13 | + | *We will participate at ‘[http://www.bio.nrw.de/studentconvention BioNRW pHD Student Convention]’ in Düsseldorf at July 13. |
Revision as of 21:52, 4 October 2013
June
Milestones
- Starting lab work on the sub-project Porins.
- Successful PCR on OprF gene from Pseudomonas fluorescens. OprF with Pre- and Suffix overlaps could be amplified from genome.
- Planning of our Human Practice projects started and the first participations are fixed.
Week 5
Organization
- iGEM-Team Bielefeld will support the ‘CeBiTec Student Academy’ from 26.-30. August with an own experiment.
MFC
Mediators
Porines
- Primer design for isolation of OprF from Pseudomonas fluorescens strain, with overlaps for BioBrick Prefix and Suffix:
- Forward Primer OprF (49 bp): GAATTCGCGGCCGCTTCTAGATGAAACTGAAAAACACCTTGGGCTTTGC
- Reverse Primer OprF (51 bp): CTGCAGCGGCCGCTACTAGTATTACTTAGCTTGGGCTTCAACCTGCGCTTC
Nanowires
- Primer design for isolation of gene-cluster GSU 1491-1495, GSU 1496-1505 and GSU Promoter-1496-1505 from Geobacter sulfurreducens, containing overlaps for BioBrick Prefix and Suffix:
- Forward GSU 1491-1495 (45 bp):
GAATTCGCGGCCGCTTCTAGATGCAGGCTAGCAGACTGGGAGAAC - Reverse GSU 1491-1495 (38 bp):
CTGCAGCGGCCGCTACTAGTATCACTCCTCATCCATGC - Forward GSU 1496-1505 (42 bp):
GAATTCGCGGCCGCTTCTAGAGTTGGCCAATTACCCCCATAC - Reverse GSU 1496-1505 (51 bp):
CTGCAGCGGCCGCTACTAGTATCATAAACGAACCTCGTCCCAAGGCATCAG - Forward GSU Promoter-1496-1505 (52 bp):
GAATTCGCGGCCGCTTCTAGAGGATAGGATCCGTCACCGAGTGCGAACTGCC
- Forward GSU 1491-1495 (45 bp):
Week 6
Organization
- Thanks to NEB for the [http://www.neb-online.de/index.php/en/neb-announcements/231-igem-2013 free iGEM kit] with many useful laboratory things for all german iGEM teams.
- We are working on our first press release.
- Having a short radio contribution in the Bielefeld university campus radio ([http://www.radiohertz.de/beta-site radio 87.9 hertz]).
MFC
Mediators
- Glycerol dehydrogenase
- Isolation of shipping vector pSB1C3 out of 2013 Distribution Kit Plate 5 Well 3A with insert Part RFP (<bbpart>J04450</bbpart>) for better transformation characterization ([http://parts.igem.org/Help:2013_DNA_Distribution Distribution Kit BioBrick isolation]).
- Transformation of <partinfo>BBa_J04450</partinfo> into Escherichia coli KRX strain.
- Plasmid isolation of <partinfo>BBa_J04450</partinfo>.
Week 7
MFC
Mediators
Cytochromes
- Cultivation of Shewanella oneidensis MR-1 in liquid LB medium at 30 °C
- Isolation of genomic DNA from S. oneidensis and dilution to the subsequently used PCR template:
- 4-2006-453: 5.5ng/µl
- Amplification of the mtrCAB cluster with Phusion polymerase
- Annealing: Gradient [55.8 - 56.7 - 57.8 - 59.1 - 60.4 - 61.7 - 62.9 - 63.9]
- Elongation: 1:15 min
- Notes: Clear Bands at the expected 5.2 kb, the annealing temperature seems not to have an effect.
- PCR-CleanUp
- Lane2: 4-2106-451: 7.4 ng/µl
- Lane5: 4-2106-451: 8.5 ng/µl
Porines
- Starting first cultivation of Pseudomonas fluorescens strain for complete genome isolation.
- Successful genome isolation of Pseudomonas fluorescens.
- Successful PCR with Forward and Reverse Primer OprF on the OprF gene of Pseudomonas fluorescens strain.
- OprF PCR product was isolated by agarose gel electrophoresis and purified.
- Bands are at expected size of 1300 bp.
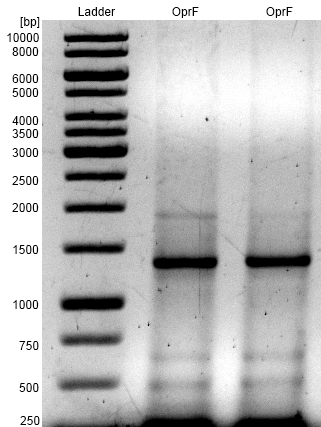
Figure 1: Agarose gel from PCR on the OprF gene of Pseudomonas fluorescens strain with forward and reverse primer OprF. For Ladder we used [http://www.thermoscientificbio.com/nucleic-acid-electrophoresis/generuler-1-kb-dna-ladder-ready-to-use-250-to-10000-bp GeneRuler™ 1 kb DNA Ladder from Thermo Scientific].
Nanowires
- Anaerobic cultivation of Geobacter sulfurreducens strain DSM-12127 in nitrogen-gassed Geobacter-medium, which was suggested by the strain-supplier: German Collection of Microorganisms and Cell Cultures DSMZ, using 30 mL cultivation-tubes and silicone stoppers with upending rim.
Week 8
Organization
- Let’s go to Lyon, flights are booked for the European jamboree in Lyon from October 11 to 13, 2013.
- We will participate at ‘[http://www.bio.nrw.de/studentconvention BioNRW pHD Student Convention]’ in Düsseldorf at July 13.
MFC
Mediators
- Glycerol dehydrogenase
- Cloning of GldA into pSB1C3 shipping vector with NEB Biobrick assembly Kit did not work as expected.
- Screening of colonies with colony PCR and Plasmid restriction analysis shows religated pSB1C3 shipping vector.
- Bands are at size of 2000 bp, the length of linear pSB1C3.
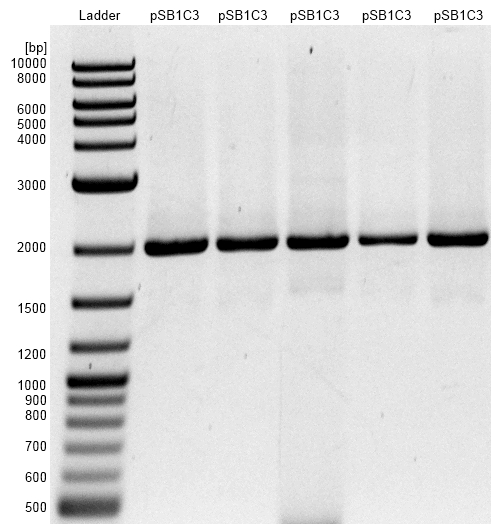
Figure 2: Agarosegel with NEB 1 kb DNA Ladder as marker. Bands are showing restriction analysis from cloning of GldA into pSB1C3 shipping vector with NEB Biobrick assembly Kit. Assembly did not work, only one band at the size of 2000 bp showing religated pSB1C3.
- Primerdesign for pSB1C3 according an universal usable backbone for Gibson Assembly with Prefix and Suffix specific overlaps:
- Forward Primer pSB1C3 (23 bp): TACTAGTAGCGGCCGCTGCAGTC
- Reverse Primer pSB1C3 (23 bp): CTCTAGAAGCGGCCGCGAATTCC
- Primerdesign for pSB1C3 according an universal usable backbone for Gibson Assembly with Prefix and Suffix specific overlaps:
Porines
- Cloning of OprF into pSB1C3 shipping vector with NEB Biobrick assembly Kit did not work as expected.
- Screening of colonies with colony PCR and Plasmid restriction analysis shows religated pSB1C3 shipping vector as described for GldA cloning.
Nanowires
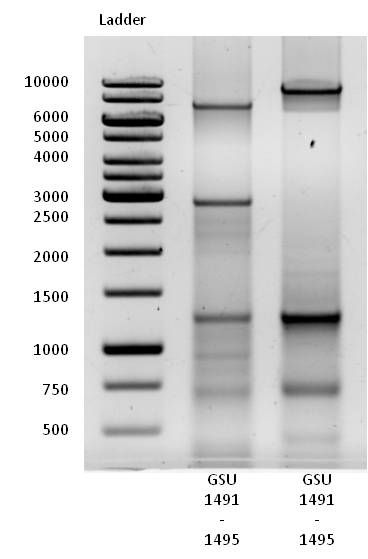
- Successful genomic DNA isolation of Geobacter sulfurreducens strain.
- Successful PCR with Forward and Reverse Primer GSU 1491-1495 and GSU 1496-1505 on the appropriate gene-cluster of Geobacter sulfurreducens.
| Component | Volume |
| H2O | to 50 µL |
| Template (100 ng) | 20 µL |
| Betain 5 M | 12.5 µL |
| 5 x Phusion GC Buffer | 10 µL |
| DMSO | 2.5 µL |
| Primer-Mix 10 mM | 2.5 µL |
| 10mM dNTP´s | 2 µL |
| Phusion DNA Polymerase | 0.5 µL |
| Step | Temperature | Time | Cycles |
| Denaturation | 98 °C | 30 sec | |
| Denaturation | 98 °C | 30 sec | 10 |
| Annealing | 55 °C | 60 sec | 10 |
| Elongation | 72°C | 270 sec | 10 |
| Denaturation | 98 °C | 30 sec | 25 |
| Annealing | 60 °C | 60 sec | 25 |
| Elongation | 72 °C | 270 sec | 25 |
| Elongation | 72 °C | 300 sec |
- Clearly visible band at size of 7200 bp, the length of gene cluster GSU 1491-1495.
- Clearly visible band at size of 9000 bp, the length of gene cluster GSU 1496-1505.
- Several bands at smaller sizes suggest alternative primer binding sites in the genome of Geobacter sulfurreducens.
- GSU 1491-1495 and GSU 1496-1505 PCR products were isolated and purified by gel extraction.
- Inoculation of new cultivation tubes, containing Geobacter-medium with Geobacter sulfurreducens culture for higher concentrated PCR-template production by whole genome isolation. Cultivation at 35 °C on a rotary shaker with 110 rpm.
 "
"