Team:Calgary/Project/OurSensor/Reporter/BetaLactamase
From 2013.igem.org
| Line 60: | Line 60: | ||
<p> As a preliminary test to confirm proper protein expression, we tested purified β-lactamase with benzylpenicillin, a substrate that gives a colourimetric and a pH output. First, we wanted to demonstrate that our bacteria carrying the <i>amp</i>R gene was expressing functional β-lactamase. <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189007">BBa_K1189007</a>. In order to do so, we performed an <a href="https://2013.igem.org/Team:Calgary/Notebook/Protocols/AmpicillinSurvivalAssay1"> | <p> As a preliminary test to confirm proper protein expression, we tested purified β-lactamase with benzylpenicillin, a substrate that gives a colourimetric and a pH output. First, we wanted to demonstrate that our bacteria carrying the <i>amp</i>R gene was expressing functional β-lactamase. <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189007">BBa_K1189007</a>. In order to do so, we performed an <a href="https://2013.igem.org/Team:Calgary/Notebook/Protocols/AmpicillinSurvivalAssay1"> | ||
ampicillin survival assay | ampicillin survival assay | ||
| - | </a> using <i>E. coli</i> transformed with a plasmid encoding the <i>amp</i>R gene. This assay would involve culturing the bacteria and then exposing them to ampcillin, and survival was then measured by OD. This then allowed us to determine whether the β-lactamase was produced and whether it is functional. Only the bacteria producing functional β-lactamase enzymes were able to survive in the presence of ampicillin resulting in an increase in OD. Whereas bacteria lacking the abililty to produce functional β-lactamase enzyme were unable to survive, seen by a decrease in OD. (Figure 6).</p> | + | </a> using <i>E. coli</i> transformed with a plasmid encoding the <i>amp</i>R gene. This assay would involve culturing the bacteria and then exposing them to ampcillin, and survival was then measured by OD. This then allowed us to determine whether the β-lactamase was produced and whether it is functional. Only the bacteria producing functional β-lactamase enzymes were able to survive in the presence of ampicillin resulting in an increase in OD. Whereas bacteria lacking the abililty to produce functional β-lactamase enzyme were unable to survive, seen by a decrease in OD. (Figure 6). Therefore, we are able to produce functional β-lactamase enzyme (Figure 6).</p> |
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2013/0/03/YYC2013_Blac_Amp_Survival_Assay_with_colonies.jpg"> | <img src="https://static.igem.org/mediawiki/2013/0/03/YYC2013_Blac_Amp_Survival_Assay_with_colonies.jpg"> | ||
| Line 66: | Line 66: | ||
<p><b>Figure 6. </b>Absorbance values at 600nm for each tube at four different time points: 0, 30, 60 and 120min. The cultures that expressed β-lactamase (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189007">BBa_K1189007</a>) showed higher absorbance levels, showing that the cells were able to grow in the presence of ampicillin.</a> | <p><b>Figure 6. </b>Absorbance values at 600nm for each tube at four different time points: 0, 30, 60 and 120min. The cultures that expressed β-lactamase (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189007">BBa_K1189007</a>) showed higher absorbance levels, showing that the cells were able to grow in the presence of ampicillin.</a> | ||
</figcaption> | </figcaption> | ||
| - | <p>After confirming protein expresison, we were able to purify both our β-lactamase (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189007">BBa_K1189007</a>) and our TALE-A-β-lactamase protein (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) (Figure 7). | + | <p>After confirming protein expresison, we wanted to demonstrate that we can also purify our proteins. Figure 7 demonstrates that we were able to purify both our β-lactamase (<a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189007">BBa_K1189007</a>) and our TALE-A-β-lactamase protein (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) (Figure 7). After purification, we wanted to demonstrate whether our purified TALE A - β-lactamase fusion protein retained its enzymatic activity. This was tested by a variation of the ampicillin survival assay where we pretreated, LB containing ampicillin and chloramphenicol, with our purified TALE A linked to β-lactamase (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>). We then cultured bacteria in the treated LB carrying the psB1C3 (<a href="http://parts.igem.org/Part:pSB1C3">pSB1C3</a>), conveying resistance to chloramphenicol. In order for the bacteria to survive our isolated protein needed to retain its enzymatic abilities. We can show that the bacteria susceptible to ampicillin were able to grow in the presence of our purified proteins (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>), which means that we are expressing and purifying functional fusion protein which is degrading the ampicillin (Figures 8). Both graphs show an increase in OD for cultures pre-treated with our protein demonstrating our protein is functional.</p> |
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2013/4/45/YYC2013_TALE_September_22_Blac.jpg"> | <img src="https://static.igem.org/mediawiki/2013/4/45/YYC2013_TALE_September_22_Blac.jpg"> | ||
| Line 88: | Line 88: | ||
<p>After verifying that <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K782004">TALE A</a>-linker-β-lactamase (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031"> | <p>After verifying that <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K782004">TALE A</a>-linker-β-lactamase (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031"> | ||
BBa_K1189031</a>) retained enzymatic activity and was able to degrade ampicillin, we performed a <a href="https://2013.igem.org/Team:Calgary/Notebook/Protocols/BenzylpenicillianAssay"> | BBa_K1189031</a>) retained enzymatic activity and was able to degrade ampicillin, we performed a <a href="https://2013.igem.org/Team:Calgary/Notebook/Protocols/BenzylpenicillianAssay"> | ||
| - | pH assay</a> using benzylpenicillin as our substrate. We were able to see a colour change due to the presence of phenol red, a pH indicator with a transition pH of 6.8-8.2, turning red at lower pH. | + | pH assay</a> using benzylpenicillin as our substrate to demonstrate that our fusion TALE A - β-lactamase protein can act as a reporter. β-lactamase hydrolyzes benzylpenicillin to penicillinoic acid, which changes the pH of the solution from alkaline to acidic. We tested it out with two separate pH indicators, phenol red and bromothymol blue, and we were able to show two different colourimetric outputs. We were able to show successful reporter acitivy with both indicators. </p> |
| + | <p> | ||
| + | We were able to see a colour change due to the presence of phenol red, a pH indicator with a transition pH of 6.8-8.2, turning red at lower pH. This pH change causes the phenol red to change from red to yellow. Our negative controls, to which benzylpenicillin was not added, remained red. We can also see the colour change coincides with the amount of purified TALE A-β-lactamase present in each sample (Figure 10). Additionally, we have demonstrated the same pH change to cause the bromothymol blue to change in colour from blue to yellow as it gets more acidic (Figure 11 & 12). We have demonstrated the reporter activity both qualitatively (Figure 10 & 11) and quantitatively (Figure 12) </p> | ||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2013/8/86/YYC2013_Blac_%2B_Penicillium_G.jpg"> | <img src="https://static.igem.org/mediawiki/2013/8/86/YYC2013_Blac_%2B_Penicillium_G.jpg"> | ||
| Line 98: | Line 100: | ||
<img src="https://static.igem.org/mediawiki/2013/c/c7/UCalgary2013TRBetalactamasecolourpsd.png" alt="Beta-lactamase Visual Assay" width="432" height="599"> | <img src="https://static.igem.org/mediawiki/2013/c/c7/UCalgary2013TRBetalactamasecolourpsd.png" alt="Beta-lactamase Visual Assay" width="432" height="599"> | ||
<figcaption> | <figcaption> | ||
| - | <p><b>Figure | + | <p><b>Figure 11.</b>Change in pH catalyzed by TALE A linked to β-lactamase (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) using benzylpenicillin. Bromothymol blue was used to keep track of this colour change. Absorbance readings were taken at 616 nm every 30 seconds. Different amounts of TALE A linked to β-lactamase (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) were tested. Commercial β-lactamase was used as a positive control. Negative controls included were bovine serum albumin, β-lactamase without the substrate and the substrate by itself.</p> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| Line 105: | Line 107: | ||
<img src="https://static.igem.org/mediawiki/2013/6/68/UCalgary2013TRBetalactamaseassay.png" alt="Beta-lactamase Catalytic Assay" width="800" height="442"> | <img src="https://static.igem.org/mediawiki/2013/6/68/UCalgary2013TRBetalactamaseassay.png" alt="Beta-lactamase Catalytic Assay" width="800" height="442"> | ||
<figcaption> | <figcaption> | ||
| - | <p><b>Figure | + | <p><b>Figure 12.</b>Change in pH catalyzed by TALE A linked to β-lactamase (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) using benzylpenicillin. Bromothymol blue was used to keep track of this colour change. Absorbance readings were taken at 616 nm every 30 seconds. Different amounts of TALE A linked to β-lactamase (<a href=" http://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) were tested. Commercial β-lactamase was used as a positive control. Negative controls included were bovine serum albumin, β-lactamase without the substrate and the substrate by itself.</p> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
Revision as of 00:41, 29 October 2013
β-Lactamase
β-Lactamase
What is β-lactamase?
β-lactamase is an enzyme encoded by the ampicillin resistance gene (ampR) frequently present in plasmids for selection. Structurally, β-lactamase is a 29 kDa monomeric enzyme (Figure 1). Its enzymatic activity provides resistance to β-lactam antibiotics such as carbapenems, penicillin and ampicillin through hydrolysis of the β-lactam ring, a structure shared by the β-lactam class of antibiotics (Qureshi, 2007).

Figure 1. 3D structure of β-lactamase obtained from our team’s work in Autodesk Maya. To learn more about our modeling, click here.
Many advantages come from working with β-lactamase. It shows high catalytic efficiency and simple kinetics. Also, no orthologs of ampR are known to be encoded by eukaryotic cells and no toxicity was identified making this protein very useful in studies involved eukaryotes (Qureshi, 2007). β-lactamase has been used to track pathogens in infected murine models (Kong et al., 2010). However, in addition to its application in eukaryotic cells,β-lactamase has been found to have an alternative application in synthetic proteins as well. β-lactamase is able to preserve its activity when fused to other proteins, meaning it can viably be used in fusion proteins (Moore et al., 1997). This feature makes β-lactamase a potentially valuable tool for assembly of synthetic constructs.
How is β-lactamase used as a Reporter?
β-lactamase, in the presence of different substrates, can give various outputs. It can produce a fluorogenic output in the presence of a cephalosporin derivative (CCF2/AM), which can then subsequently be measured using a fluorometer (Remy et al., 2007). Additionally, β-lactamase can also be used to obtain colourimetric outputs by breaking down synthetic compounds such as nitrocefin (Figure 2). The result of nitrocefin hydrolysis is a colour change from yellow to red(Remy et al., 2007). A third output that β-lactamase can give out is through pH. One example is the hydrolysis of benzylpenicillin by β-lactamase, converting the substrate to an acid and lowering pH. This can then be seen through the use of pH indicators such as phenol red to give an observable output (Li et al., 2008). The multiple ways this enzyme can be used shows the versatillity of it, as it is capable of three different outputs, fluorescent, colourimetric, and pH.

Figure 2. Hydrolysis of nitrocefin catalyzed β-lactamase, which causes a colour change from yellow to red.
β-lactamase can also be split apart in to two halves for protein complementation assays, where each half is linked to one of the two proteins being tested. If the two proteins interact the two halves are able to fold into their correct structure and give an output (Wehrman et al., 2002).
Therefore, this enzyme gives a lot of flexibility, both in how it can be attached to proteins as well as the various outputs it can give, making it a useful reporter to characterize and add to the Parts Registry.
How does β-lactamase fit in our Biosensor?
β-lactamase serves as another reporter we explored for our system in parallel to our Prussian Prussian blue ferritin reporter. But unlike the Prussian Blue ferritin system, β-lactamase can also be used for a pH output as well. This would add more versatility to our system, as β-lactamase can add a different type of output which would mean that our system is not only limited to the colourimetric outputs. Additionally, the pH output can be paired with different pH indicators to give a variety of colourimetric outputs. We have characterized this ability with two separate pH indicators, phenol red and bromothymol blue, and were able to demonstrate two completely different colourimetric output. In addition, the output of our system can be scaled by altering the number of fused β-lactamase proteins by exploiting the ferritin nanoparticle. This can be achieved through modifying the number of β-lactamase molecules attached to ferritin, ranging from 24 or 12 depending on whether our ferritin nanoparticle consists of the 12 heavy-light subunit fusions (BBa_K157018),or 24 individual subunits, composed of separate light and heavy subunits. The result is a system that can be scaled by utilizing 24 or 12 β-lactamase proteins, or only 1 Prussian blue ferritin core, or one β-lactamase protein fused to a TALE as an alternate. This adds the elements of versatility, flexibility, and most importantly, modularity to our biosensor and serves a vital basis for our platform.
Constructs
We retrieved the ampR gene from the backbone of the pSB1A3 plasmid. We added a a His-tag was added to the N-terminus of it using a flexible glycine linker (BBa_K157013), allowing purification through Ni-NTA protein purification, as well as the lacI promoter for expression (Figure 3). Additionaly, we modified this gene to make it a more useful part for the registry, such as the removal of a BsaI cut-site, making it viable for Golden Gate assembly (Figure 4). We also fused the ampR gene to on our TALE A to bind to our target sequence (Figure 5). This could be used in conjunction with another TALE to act in our strip assay. These modifications resulted in the products shown below:

Figure 3. On the left, part BBa_K1189009. We added a His-tag to β-lactamase to facilitate purification. On the right, part BBa_K1189007. In addition to the His-tag, PLacI + RBS were added upstream of the β-lactamase gene so we can express and characterize our part.

Figure 4. Part BBa_K1189008. We removed the BsaI cut site in the β-lactamase gene so that it could be used for Golden Gate Assembly.

Figure 5. Part BBa_K1189031. This construct works as the mobile detector in our biosensor. TALE A is linked to β-lactamase and if the stx2 gene is present in the strip, our mobile is retained on the strip so β-lactamase can give a colour output in the presence of a substrate.
Results
As a preliminary test to confirm proper protein expression, we tested purified β-lactamase with benzylpenicillin, a substrate that gives a colourimetric and a pH output. First, we wanted to demonstrate that our bacteria carrying the ampR gene was expressing functional β-lactamase. BBa_K1189007. In order to do so, we performed an ampicillin survival assay using E. coli transformed with a plasmid encoding the ampR gene. This assay would involve culturing the bacteria and then exposing them to ampcillin, and survival was then measured by OD. This then allowed us to determine whether the β-lactamase was produced and whether it is functional. Only the bacteria producing functional β-lactamase enzymes were able to survive in the presence of ampicillin resulting in an increase in OD. Whereas bacteria lacking the abililty to produce functional β-lactamase enzyme were unable to survive, seen by a decrease in OD. (Figure 6). Therefore, we are able to produce functional β-lactamase enzyme (Figure 6).

Figure 6. Absorbance values at 600nm for each tube at four different time points: 0, 30, 60 and 120min. The cultures that expressed β-lactamase (BBa_K1189007) showed higher absorbance levels, showing that the cells were able to grow in the presence of ampicillin.
After confirming protein expresison, we wanted to demonstrate that we can also purify our proteins. Figure 7 demonstrates that we were able to purify both our β-lactamase (BBa_K1189007) and our TALE-A-β-lactamase protein (BBa_K1189031) (Figure 7). After purification, we wanted to demonstrate whether our purified TALE A - β-lactamase fusion protein retained its enzymatic activity. This was tested by a variation of the ampicillin survival assay where we pretreated, LB containing ampicillin and chloramphenicol, with our purified TALE A linked to β-lactamase (BBa_K1189031). We then cultured bacteria in the treated LB carrying the psB1C3 (pSB1C3), conveying resistance to chloramphenicol. In order for the bacteria to survive our isolated protein needed to retain its enzymatic abilities. We can show that the bacteria susceptible to ampicillin were able to grow in the presence of our purified proteins (BBa_K1189031), which means that we are expressing and purifying functional fusion protein which is degrading the ampicillin (Figures 8). Both graphs show an increase in OD for cultures pre-treated with our protein demonstrating our protein is functional.

Figure 7. On the left crude lysate of β-lactamase + His (BBa_K1189007) from different lysis protocols, beat beating and sucrose osmotic shock respectively. On the right, western blot of TALE A-linker-β-lactamase (BBa_K1189031) showing that we were able to express and purify our fusion protein.

Figure 8. Absorbance values at 600nm after 24h. Amount of protein added ranged from 0.1µg to 20µg of TALE A-link-β-lactamase (BBa_K1189031) were sufficient to degrade the ampicillin in the media allowing bacteria susceptible to ampicillin to grow.

Figure 9. Absorbance values at 600nm in different time points. Amounts from 1.0µg to 10µg of TALE A-link-β-lactamase (BBa_K1189031) were sufficient to degrade the ampicillin in the media allowing bacteria susceptible to ampicillin to grow.
After verifying that TALE A-linker-β-lactamase ( BBa_K1189031) retained enzymatic activity and was able to degrade ampicillin, we performed a pH assay using benzylpenicillin as our substrate to demonstrate that our fusion TALE A - β-lactamase protein can act as a reporter. β-lactamase hydrolyzes benzylpenicillin to penicillinoic acid, which changes the pH of the solution from alkaline to acidic. We tested it out with two separate pH indicators, phenol red and bromothymol blue, and we were able to show two different colourimetric outputs. We were able to show successful reporter acitivy with both indicators.
We were able to see a colour change due to the presence of phenol red, a pH indicator with a transition pH of 6.8-8.2, turning red at lower pH. This pH change causes the phenol red to change from red to yellow. Our negative controls, to which benzylpenicillin was not added, remained red. We can also see the colour change coincides with the amount of purified TALE A-β-lactamase present in each sample (Figure 10). Additionally, we have demonstrated the same pH change to cause the bromothymol blue to change in colour from blue to yellow as it gets more acidic (Figure 11 & 12). We have demonstrated the reporter activity both qualitatively (Figure 10 & 11) and quantitatively (Figure 12)

Figure 10. Benzylpenicillin assay. On the top, the wells only had TALE A-linker-β-lactamase (BBa_K1189031). Benzylpenicillin was added and after a 10-minute incubation at room temperature, we were able to observe a colour output from red to yellow (bottom row) while the control wells remained red.

Figure 11.Change in pH catalyzed by TALE A linked to β-lactamase (BBa_K1189031) using benzylpenicillin. Bromothymol blue was used to keep track of this colour change. Absorbance readings were taken at 616 nm every 30 seconds. Different amounts of TALE A linked to β-lactamase (BBa_K1189031) were tested. Commercial β-lactamase was used as a positive control. Negative controls included were bovine serum albumin, β-lactamase without the substrate and the substrate by itself.
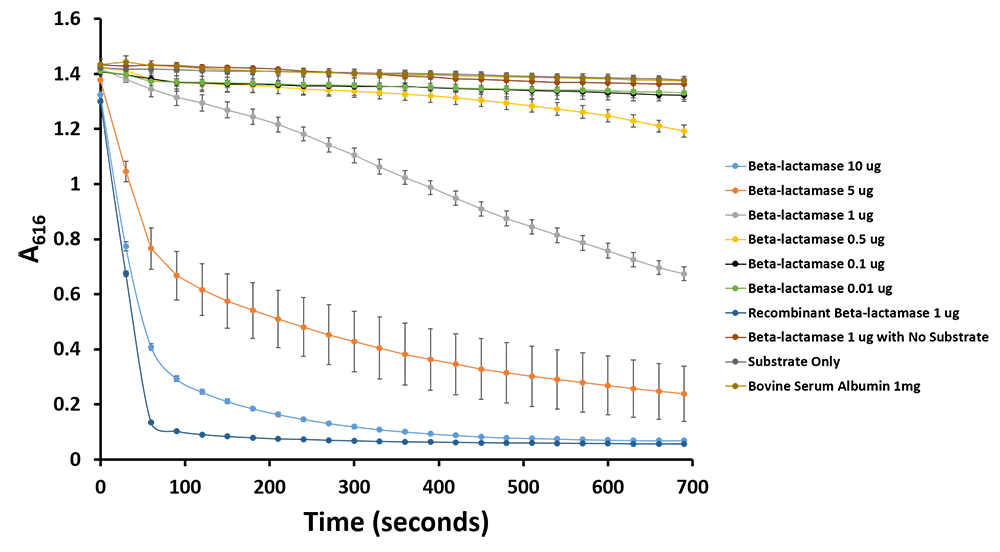
Figure 12.Change in pH catalyzed by TALE A linked to β-lactamase (BBa_K1189031) using benzylpenicillin. Bromothymol blue was used to keep track of this colour change. Absorbance readings were taken at 616 nm every 30 seconds. Different amounts of TALE A linked to β-lactamase (BBa_K1189031) were tested. Commercial β-lactamase was used as a positive control. Negative controls included were bovine serum albumin, β-lactamase without the substrate and the substrate by itself.
Therefore, we have built and submitted parts containing β-lactamase, both on its own and linked to TALE A. We then expressed, purified and demonstrated the final purified products have retained their enzymatic activity. We can show activity for our mobile TALE A linked to β-lactamase (BBa_K1189031) for our sensor in two different ways, through pH and cell growth assays.
 "
"
