Team:Evry/LogisticFunctions
From 2013.igem.org
(Difference between revisions)
| Line 14: | Line 14: | ||
<h2>Parameters:</h2> | <h2>Parameters:</h2> | ||
<ul> | <ul> | ||
| - | <li><b>Q</b> : Magnitude.<br/>lim(g)=Q</li> | + | <li><b>Q</b> : Magnitude.<br/> <i>lim(g)=Q</i></li> |
<li><b>d</b> : Offset.</li> | <li><b>d</b> : Offset.</li> | ||
<li><b>p</b> : Precision.<br/>g(d)=Q*p</li> | <li><b>p</b> : Precision.<br/>g(d)=Q*p</li> | ||
Revision as of 10:45, 29 August 2013
Logistic functions :
When we started to model biological behaviors, we realised very soon that we were going to need a function that simulates a non-exponential evolution, that would include a simple speed control and a maximum value. A smooth step function.
Such functions, named logistic functions were introduced around 1840 by M. Verhulst.
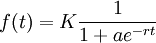
These functions looked perfect, but we needed more control : we needed to set a starting value and a precision.

Parameters:
- Q : Magnitude.
lim(g)=Q - d : Offset.
- p : Precision.
g(d)=Q*p - k : Efficiency.

References:
 "
"













