Team:TU-Munich/Results/Recombinant
From 2013.igem.org
(Difference between revisions)
(→Characterization of recombinant effector proteins) |
|||
| Line 6: | Line 6: | ||
<!-- Start of content --> | <!-- Start of content --> | ||
| - | |||
==Characterization of recombinant effector proteins== | ==Characterization of recombinant effector proteins== | ||
Text. See [https://2013.igem.org/Team:TU-Munich/Notebook/Methods proteinbiochemical methods] for further informatons. | Text. See [https://2013.igem.org/Team:TU-Munich/Notebook/Methods proteinbiochemical methods] for further informatons. | ||
{|cellspacing="0" border="1" right | {|cellspacing="0" border="1" right | ||
| - | | | + | |+ Table 1: Investigated Proteins |
| - | + | !Protein | |
| - | + | !BioBrick | |
| - | + | !RFC | |
| - | + | !Affinity tag | |
| - | + | !Size [kDa] | |
| - | + | !Disulphid bridges | |
| - | + | !comment | |
| - | + | ||
| - | + | ||
|- | |- | ||
|Eryhtromycin esterase (EreB) | |Eryhtromycin esterase (EreB) | ||
| Line 74: | Line 71: | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
==Eryhtromycin Esterase== | ==Eryhtromycin Esterase== | ||
| Line 97: | Line 90: | ||
<br><br> | <br><br> | ||
| - | |||
| - | |||
==Laccase== | ==Laccase== | ||
| Line 108: | Line 99: | ||
<html><!--- Please copy this table containing parameters for BBa_ at the end of the parametrs section ahead of the references. ---><style type="text/css">table#AutoAnnotator {border:1px solid black; width:100%; border-collapse:collapse;} th#AutoAnnotatorHeader { border:1px solid black; width:100%; background-color: rgb(221, 221, 221);} td.AutoAnnotator1col { width:100%; border:1px solid black; } span.AutoAnnotatorSequence { font-family:'Courier New', Arial; } td.AutoAnnotatorSeqNum { text-align:right; width:2%; } td.AutoAnnotatorSeqSeq { width:98% } td.AutoAnnotatorSeqFeat1 { width:3% } td.AutoAnnotatorSeqFeat2a { width:27% } td.AutoAnnotatorSeqFeat2b { width:97% } td.AutoAnnotatorSeqFeat3 { width:70% } table.AutoAnnotatorNoBorder { border:0px; width:100%; border-collapse:collapse; } table.AutoAnnotatorWithBorder { border:1px solid black; width:100%; border-collapse:collapse; } td.AutoAnnotatorOuterAmino { border:0px solid black; width:20% } td.AutoAnnotatorInnerAmino { border:1px solid black; width:50% } td.AutoAnnotatorAminoCountingOuter { border:1px solid black; width:40%; } td.AutoAnnotatorBiochemParOuter { border:1px solid black; width:60%; } td.AutoAnnotatorAminoCountingInner1 { width: 7.5% } td.AutoAnnotatorAminoCountingInner2 { width:62.5% } td.AutoAnnotatorAminoCountingInner3 { width:30% } td.AutoAnnotatorBiochemParInner1 { width: 5% } td.AutoAnnotatorBiochemParInner2 { width:55% } td.AutoAnnotatorBiochemParInner3 { width:40% } td.AutoAnnotatorCodonUsage1 { width: 3% } td.AutoAnnotatorCodonUsage2 { width:14.2% } td.AutoAnnotatorCodonUsage3 { width:13.8% } </style><table id="AutoAnnotator"><tr><!-- Time stamp in ms since 1/1/1970 1379191798681 --><th id="AutoAnnotatorHeader" colspan="2">Protein data table for BioBrick <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_<!------------------------Enter BioBrick number here------------------------>">BBa_<!------------------------Enter BioBrick number here------------------------></a> automatically created by the <a href="https://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">BioBrick-AutoAnnotator</a> version 1.0</th></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Nucleotide sequence</strong> in <strong>RFC 25</strong>, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein)<br><span class="AutoAnnotatorSequence"> <u><i>ATGGCCGGC</i>AACCTAGAA ... GATATCATC<i>ACCGGT</i></u></span><br> <strong>ORF</strong> from nucleotide position -8 to 1530 (excluding stop-codon)</td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid sequence:</strong> (RFC25 scars in shown in bold, other sequence features underlined; both given below)<br><span class="AutoAnnotatorSequence"><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqNum">1 </td><td class="AutoAnnotatorSeqSeq">MAGNLEKFVDELPIPEVAKPVKKNPKQTYYEIAMEEVFLKVHRDLPPTKLWTYNGSLPGPTIHANRNEKVKVKWMNKLPLKHFLPVDHTIHEGHHDEPEV</td></tr><tr><td class="AutoAnnotatorSeqNum">101 </td><td class="AutoAnnotatorSeqSeq">KTVVHLHGGVTPASSDGYPEAWFSRDFEA<b>TG</b>PFFEREVYEYPNHQQACTLWYHDHAMALTRLNVYAGLAGFYLISDAFEKSLELPKGEYDIPLMIMDRTF</td></tr><tr><td class="AutoAnnotatorSeqNum">201 </td><td class="AutoAnnotatorSeqSeq">QEDGALFYPSRPNNTPEDSDIPDPSIVPFFCGETILVNGKVWPYLEVEPRKYRFRILNASNTRTYELHLDNDATILQIGSDGGFLPRPVHHQSFSIAPAE</td></tr><tr><td class="AutoAnnotatorSeqNum">301 </td><td class="AutoAnnotatorSeqSeq">RFDVIIDFSAYENKTITLKNKAGCGQEVNPETDANIMQFKVTRPLKGRAPKTLRPIFKPLPPLRPCRADKERTLTLTGTQDKYGRPILLLDNQFWNDPVT</td></tr><tr><td class="AutoAnnotatorSeqNum">401 </td><td class="AutoAnnotatorSeqSeq">ENPRLGSVEVWSIVNPTRGTHPIHLHLVQFRVIDRRPFDTEVYQSTGDIVYTGPNEAPPLHEQGYKDTIQAHAGEVIRIIARFVPYSGRYVWHCHILEHE</td></tr><tr><td class="AutoAnnotatorSeqNum">501 </td><td class="AutoAnnotatorSeqSeq">DYDMMRPMDIITG*</td></tr></table></span></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Sequence features:</strong> (with their position in the amino acid sequence, see the <a href="https://2013.igem.org/Team:TU-Munich/Results/Software/FeatureList">list of supported features</a>)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqFeat1"></td><td class="AutoAnnotatorSeqFeat2a">RFC25 scar (shown in bold): </td><td class="AutoAnnotatorSeqFeat3">130 to 131</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid composition:</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Ala (A)</td><td class="AutoAnnotatorInnerAmino">27 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Arg (R)</td><td class="AutoAnnotatorInnerAmino">29 (5.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asn (N)</td><td class="AutoAnnotatorInnerAmino">23 (4.5%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asp (D)</td><td class="AutoAnnotatorInnerAmino">31 (6.0%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Cys (C)</td><td class="AutoAnnotatorInnerAmino">5 (1.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gln (Q)</td><td class="AutoAnnotatorInnerAmino">14 (2.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Glu (E)</td><td class="AutoAnnotatorInnerAmino">37 (7.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gly (G)</td><td class="AutoAnnotatorInnerAmino">30 (5.8%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">His (H)</td><td class="AutoAnnotatorInnerAmino">23 (4.5%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ile (I)</td><td class="AutoAnnotatorInnerAmino">31 (6.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Leu (L)</td><td class="AutoAnnotatorInnerAmino">41 (8.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Lys (K)</td><td class="AutoAnnotatorInnerAmino">27 (5.3%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Met (M)</td><td class="AutoAnnotatorInnerAmino">10 (1.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Phe (F)</td><td class="AutoAnnotatorInnerAmino">24 (4.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Pro (P)</td><td class="AutoAnnotatorInnerAmino">47 (9.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ser (S)</td><td class="AutoAnnotatorInnerAmino">18 (3.5%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Thr (T)</td><td class="AutoAnnotatorInnerAmino">33 (6.4%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Trp (W)</td><td class="AutoAnnotatorInnerAmino">8 (1.6%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Tyr (Y)</td><td class="AutoAnnotatorInnerAmino">22 (4.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Val (V)</td><td class="AutoAnnotatorInnerAmino">33 (6.4%)</td></tr></table></td></tr></table></td></tr><tr><td class="AutoAnnotatorAminoCountingOuter"><strong>Amino acid counting</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Total number:</td><td class="AutoAnnotatorAminoCountingInner3">513</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Positively charged (Arg+Lys):</td><td class="AutoAnnotatorAminoCountingInner3">56 (10.9%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Negatively charged (Asp+Glu):</td><td class="AutoAnnotatorAminoCountingInner3">68 (13.3%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Aromatic (Phe+His+Try+Tyr):</td><td class="AutoAnnotatorAminoCountingInner3">77 (15.0%)</td></tr></table></td><td class="AutoAnnotatorBiochemParOuter"><strong>Biochemical parameters</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Atomic composition:</td><td class="AutoAnnotatorBiochemParInner3">C<sub>2671</sub>H<sub>4072</sub>N<sub>718</sub>O<sub>760</sub>S<sub>15</sub></td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Molecular mass [Da]:</td><td class="AutoAnnotatorBiochemParInner3">58883.0</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Theoretical pI:</td><td class="AutoAnnotatorBiochemParInner3">6.03</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Extinction coefficient at 280 nm [M<sup>-1</sup> cm<sup>-1</sup>]:</td><td class="AutoAnnotatorBiochemParInner3">76780 / 77093 (all Cys red/ox)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Codon usage</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Organism:</td><td class="AutoAnnotatorCodonUsage3"><i>E. coli</i></td><td class="AutoAnnotatorCodonUsage3"><i>B. subtilis</i></td><td class="AutoAnnotatorCodonUsage3"><i>S. cerevisiae</i></td><td class="AutoAnnotatorCodonUsage3"><i>A. thaliana</i></td><td class="AutoAnnotatorCodonUsage3"><i>P. patens</i></td><td class="AutoAnnotatorCodonUsage3">Mammals</td></tr><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Codon quality (<a href="http://en.wikipedia.org/wiki/Codon_Adaptation_Index">CAI</a>):</td><td class="AutoAnnotatorCodonUsage3">good (0.72)</td><td class="AutoAnnotatorCodonUsage3">good (0.75)</td><td class="AutoAnnotatorCodonUsage3">good (0.68)</td><td class="AutoAnnotatorCodonUsage3">good (0.74)</td><td class="AutoAnnotatorCodonUsage3">good (0.77)</td><td class="AutoAnnotatorCodonUsage3">good (0.67)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"> The BioBrick-AutoAnnotator was created by <a href="https://2013.igem.org/Team:TU-Munich">TU-Munich 2013</a> iGEM team. For more information please see the <a href="https://2013.igem.org/Team:TU-Munich/Results/Software">documentation</a>.<br>If you have any questions, comments or suggestions, please leave us a <a href="https://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">comment</a>.</td></tr></table><br></html> | <html><!--- Please copy this table containing parameters for BBa_ at the end of the parametrs section ahead of the references. ---><style type="text/css">table#AutoAnnotator {border:1px solid black; width:100%; border-collapse:collapse;} th#AutoAnnotatorHeader { border:1px solid black; width:100%; background-color: rgb(221, 221, 221);} td.AutoAnnotator1col { width:100%; border:1px solid black; } span.AutoAnnotatorSequence { font-family:'Courier New', Arial; } td.AutoAnnotatorSeqNum { text-align:right; width:2%; } td.AutoAnnotatorSeqSeq { width:98% } td.AutoAnnotatorSeqFeat1 { width:3% } td.AutoAnnotatorSeqFeat2a { width:27% } td.AutoAnnotatorSeqFeat2b { width:97% } td.AutoAnnotatorSeqFeat3 { width:70% } table.AutoAnnotatorNoBorder { border:0px; width:100%; border-collapse:collapse; } table.AutoAnnotatorWithBorder { border:1px solid black; width:100%; border-collapse:collapse; } td.AutoAnnotatorOuterAmino { border:0px solid black; width:20% } td.AutoAnnotatorInnerAmino { border:1px solid black; width:50% } td.AutoAnnotatorAminoCountingOuter { border:1px solid black; width:40%; } td.AutoAnnotatorBiochemParOuter { border:1px solid black; width:60%; } td.AutoAnnotatorAminoCountingInner1 { width: 7.5% } td.AutoAnnotatorAminoCountingInner2 { width:62.5% } td.AutoAnnotatorAminoCountingInner3 { width:30% } td.AutoAnnotatorBiochemParInner1 { width: 5% } td.AutoAnnotatorBiochemParInner2 { width:55% } td.AutoAnnotatorBiochemParInner3 { width:40% } td.AutoAnnotatorCodonUsage1 { width: 3% } td.AutoAnnotatorCodonUsage2 { width:14.2% } td.AutoAnnotatorCodonUsage3 { width:13.8% } </style><table id="AutoAnnotator"><tr><!-- Time stamp in ms since 1/1/1970 1379191798681 --><th id="AutoAnnotatorHeader" colspan="2">Protein data table for BioBrick <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_<!------------------------Enter BioBrick number here------------------------>">BBa_<!------------------------Enter BioBrick number here------------------------></a> automatically created by the <a href="https://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">BioBrick-AutoAnnotator</a> version 1.0</th></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Nucleotide sequence</strong> in <strong>RFC 25</strong>, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein)<br><span class="AutoAnnotatorSequence"> <u><i>ATGGCCGGC</i>AACCTAGAA ... GATATCATC<i>ACCGGT</i></u></span><br> <strong>ORF</strong> from nucleotide position -8 to 1530 (excluding stop-codon)</td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid sequence:</strong> (RFC25 scars in shown in bold, other sequence features underlined; both given below)<br><span class="AutoAnnotatorSequence"><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqNum">1 </td><td class="AutoAnnotatorSeqSeq">MAGNLEKFVDELPIPEVAKPVKKNPKQTYYEIAMEEVFLKVHRDLPPTKLWTYNGSLPGPTIHANRNEKVKVKWMNKLPLKHFLPVDHTIHEGHHDEPEV</td></tr><tr><td class="AutoAnnotatorSeqNum">101 </td><td class="AutoAnnotatorSeqSeq">KTVVHLHGGVTPASSDGYPEAWFSRDFEA<b>TG</b>PFFEREVYEYPNHQQACTLWYHDHAMALTRLNVYAGLAGFYLISDAFEKSLELPKGEYDIPLMIMDRTF</td></tr><tr><td class="AutoAnnotatorSeqNum">201 </td><td class="AutoAnnotatorSeqSeq">QEDGALFYPSRPNNTPEDSDIPDPSIVPFFCGETILVNGKVWPYLEVEPRKYRFRILNASNTRTYELHLDNDATILQIGSDGGFLPRPVHHQSFSIAPAE</td></tr><tr><td class="AutoAnnotatorSeqNum">301 </td><td class="AutoAnnotatorSeqSeq">RFDVIIDFSAYENKTITLKNKAGCGQEVNPETDANIMQFKVTRPLKGRAPKTLRPIFKPLPPLRPCRADKERTLTLTGTQDKYGRPILLLDNQFWNDPVT</td></tr><tr><td class="AutoAnnotatorSeqNum">401 </td><td class="AutoAnnotatorSeqSeq">ENPRLGSVEVWSIVNPTRGTHPIHLHLVQFRVIDRRPFDTEVYQSTGDIVYTGPNEAPPLHEQGYKDTIQAHAGEVIRIIARFVPYSGRYVWHCHILEHE</td></tr><tr><td class="AutoAnnotatorSeqNum">501 </td><td class="AutoAnnotatorSeqSeq">DYDMMRPMDIITG*</td></tr></table></span></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Sequence features:</strong> (with their position in the amino acid sequence, see the <a href="https://2013.igem.org/Team:TU-Munich/Results/Software/FeatureList">list of supported features</a>)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqFeat1"></td><td class="AutoAnnotatorSeqFeat2a">RFC25 scar (shown in bold): </td><td class="AutoAnnotatorSeqFeat3">130 to 131</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid composition:</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Ala (A)</td><td class="AutoAnnotatorInnerAmino">27 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Arg (R)</td><td class="AutoAnnotatorInnerAmino">29 (5.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asn (N)</td><td class="AutoAnnotatorInnerAmino">23 (4.5%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asp (D)</td><td class="AutoAnnotatorInnerAmino">31 (6.0%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Cys (C)</td><td class="AutoAnnotatorInnerAmino">5 (1.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gln (Q)</td><td class="AutoAnnotatorInnerAmino">14 (2.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Glu (E)</td><td class="AutoAnnotatorInnerAmino">37 (7.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gly (G)</td><td class="AutoAnnotatorInnerAmino">30 (5.8%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">His (H)</td><td class="AutoAnnotatorInnerAmino">23 (4.5%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ile (I)</td><td class="AutoAnnotatorInnerAmino">31 (6.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Leu (L)</td><td class="AutoAnnotatorInnerAmino">41 (8.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Lys (K)</td><td class="AutoAnnotatorInnerAmino">27 (5.3%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Met (M)</td><td class="AutoAnnotatorInnerAmino">10 (1.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Phe (F)</td><td class="AutoAnnotatorInnerAmino">24 (4.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Pro (P)</td><td class="AutoAnnotatorInnerAmino">47 (9.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ser (S)</td><td class="AutoAnnotatorInnerAmino">18 (3.5%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Thr (T)</td><td class="AutoAnnotatorInnerAmino">33 (6.4%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Trp (W)</td><td class="AutoAnnotatorInnerAmino">8 (1.6%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Tyr (Y)</td><td class="AutoAnnotatorInnerAmino">22 (4.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Val (V)</td><td class="AutoAnnotatorInnerAmino">33 (6.4%)</td></tr></table></td></tr></table></td></tr><tr><td class="AutoAnnotatorAminoCountingOuter"><strong>Amino acid counting</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Total number:</td><td class="AutoAnnotatorAminoCountingInner3">513</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Positively charged (Arg+Lys):</td><td class="AutoAnnotatorAminoCountingInner3">56 (10.9%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Negatively charged (Asp+Glu):</td><td class="AutoAnnotatorAminoCountingInner3">68 (13.3%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Aromatic (Phe+His+Try+Tyr):</td><td class="AutoAnnotatorAminoCountingInner3">77 (15.0%)</td></tr></table></td><td class="AutoAnnotatorBiochemParOuter"><strong>Biochemical parameters</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Atomic composition:</td><td class="AutoAnnotatorBiochemParInner3">C<sub>2671</sub>H<sub>4072</sub>N<sub>718</sub>O<sub>760</sub>S<sub>15</sub></td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Molecular mass [Da]:</td><td class="AutoAnnotatorBiochemParInner3">58883.0</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Theoretical pI:</td><td class="AutoAnnotatorBiochemParInner3">6.03</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Extinction coefficient at 280 nm [M<sup>-1</sup> cm<sup>-1</sup>]:</td><td class="AutoAnnotatorBiochemParInner3">76780 / 77093 (all Cys red/ox)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Codon usage</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Organism:</td><td class="AutoAnnotatorCodonUsage3"><i>E. coli</i></td><td class="AutoAnnotatorCodonUsage3"><i>B. subtilis</i></td><td class="AutoAnnotatorCodonUsage3"><i>S. cerevisiae</i></td><td class="AutoAnnotatorCodonUsage3"><i>A. thaliana</i></td><td class="AutoAnnotatorCodonUsage3"><i>P. patens</i></td><td class="AutoAnnotatorCodonUsage3">Mammals</td></tr><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Codon quality (<a href="http://en.wikipedia.org/wiki/Codon_Adaptation_Index">CAI</a>):</td><td class="AutoAnnotatorCodonUsage3">good (0.72)</td><td class="AutoAnnotatorCodonUsage3">good (0.75)</td><td class="AutoAnnotatorCodonUsage3">good (0.68)</td><td class="AutoAnnotatorCodonUsage3">good (0.74)</td><td class="AutoAnnotatorCodonUsage3">good (0.77)</td><td class="AutoAnnotatorCodonUsage3">good (0.67)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"> The BioBrick-AutoAnnotator was created by <a href="https://2013.igem.org/Team:TU-Munich">TU-Munich 2013</a> iGEM team. For more information please see the <a href="https://2013.igem.org/Team:TU-Munich/Results/Software">documentation</a>.<br>If you have any questions, comments or suggestions, please leave us a <a href="https://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">comment</a>.</td></tr></table><br></html> | ||
| + | |||
===Laccase a is a secreted enzyme=== | ===Laccase a is a secreted enzyme=== | ||
[[File:TUM13_Laccase_DSB.png|thumb|right|320px| Figure 3:]]---> | [[File:TUM13_Laccase_DSB.png|thumb|right|320px| Figure 3:]]---> | ||
| Line 123: | Line 115: | ||
[[File:TUM13_Laccase_activity.png|thumb|right|320px| Figure 3:]] | [[File:TUM13_Laccase_activity.png|thumb|right|320px| Figure 3:]] | ||
| - | |||
| - | |||
==Nano Luciferase== | ==Nano Luciferase== | ||
| Line 143: | Line 133: | ||
| - | |||
| - | |||
| - | |||
| - | |||
==XylE== | ==XylE== | ||
| Line 158: | Line 144: | ||
<br><br> | <br><br> | ||
| - | |||
| - | |||
| - | |||
==DDT-Dehydrochlorinase== | ==DDT-Dehydrochlorinase== | ||
| Line 173: | Line 156: | ||
<br><br> | <br><br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
==PP1== | ==PP1== | ||
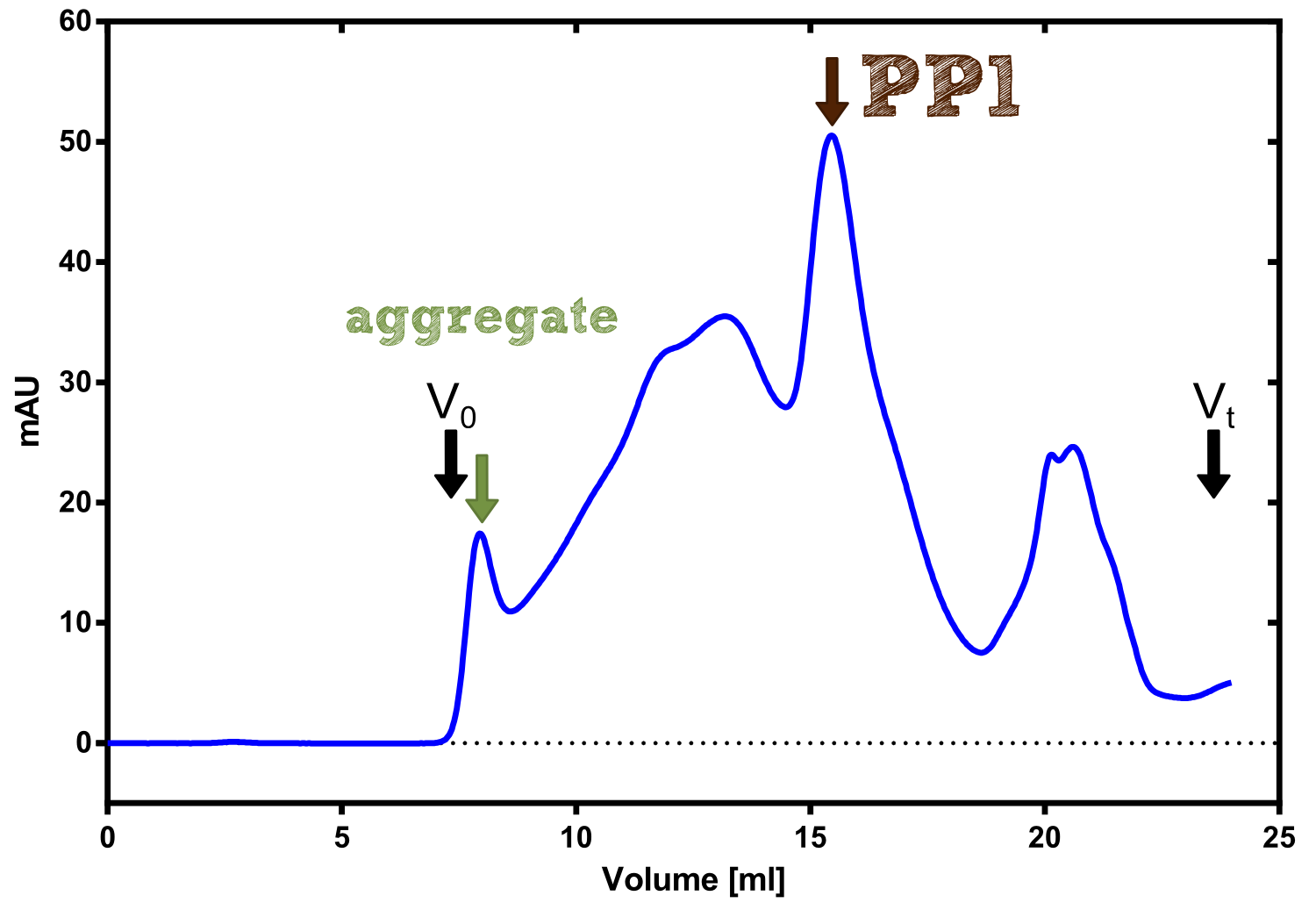
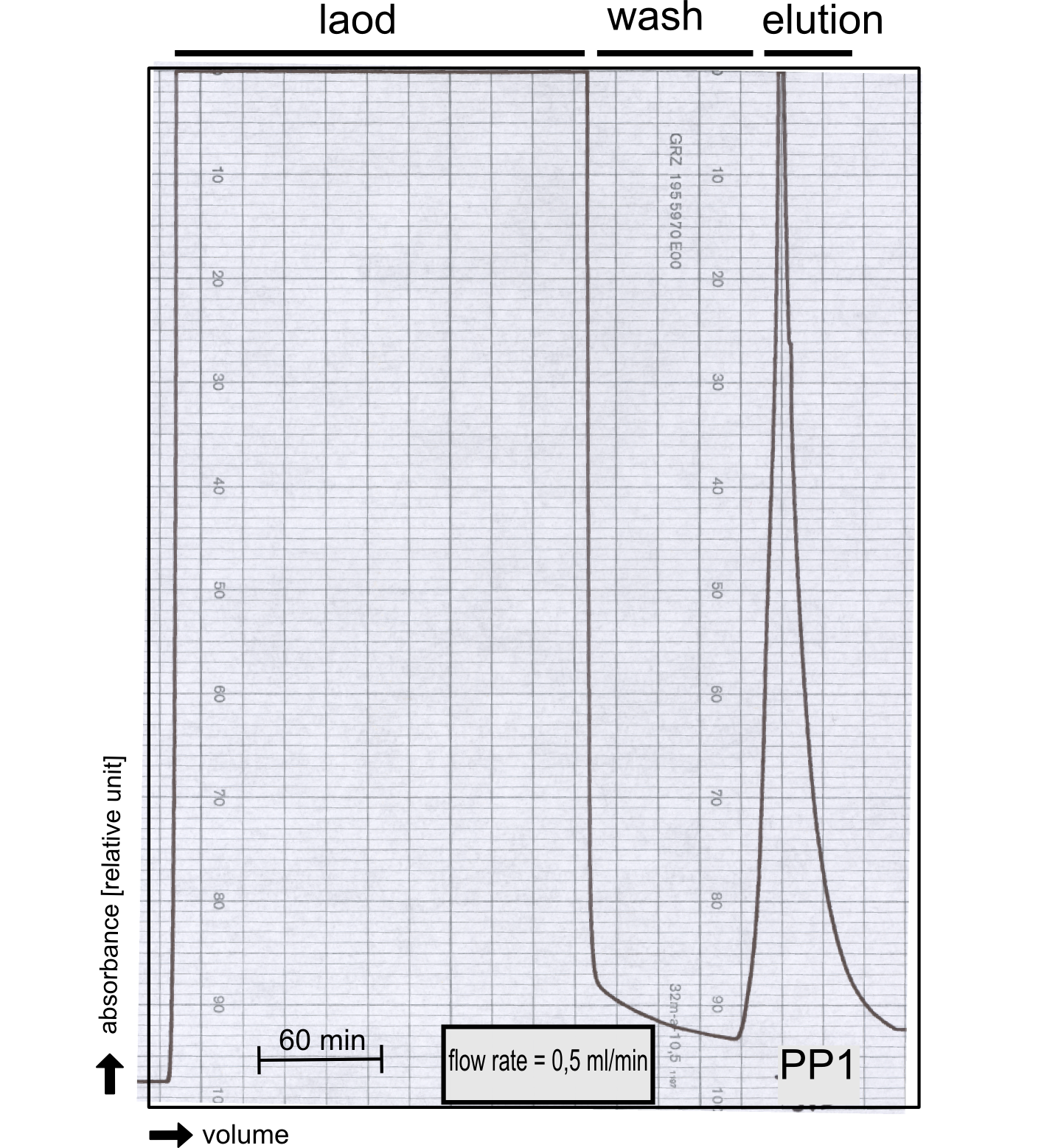
[[File:TUM13_PP1_analy_preparation.png|thumb|right|320px| Figure 3:]] | [[File:TUM13_PP1_analy_preparation.png|thumb|right|320px| Figure 3:]] | ||
[[File:TUM13_P913.png|thumb|right|320px| Figure 3:]] | [[File:TUM13_P913.png|thumb|right|320px| Figure 3:]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
==SpyCatcher & SpyTag== | ==SpyCatcher & SpyTag== | ||
| Line 200: | Line 175: | ||
[...] Characterization | [...] Characterization | ||
<br><br> | <br><br> | ||
| - | + | ||
==References:== | ==References:== | ||
| - | + | ||
<!-- Kopiervorlagen --> | <!-- Kopiervorlagen --> | ||
[[http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984]] | [[http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984]] | ||
| Line 209: | Line 184: | ||
<!-- Ab hier richtige Referenzen einfügen --> | <!-- Ab hier richtige Referenzen einfügen --> | ||
#[[http://udel.edu/~gshriver/pdf/Pimenteletal1997.pdf Pmentel et al., 1997]] Pimentel, D., Wilson, C., McCullum, C., Huang, R., Dwen, P., Flack, J. Tran, Q., Saltman, T., Cliff, T. (1997). Economic and environmental benefits of biodiversity. ''BioScience'', Vol. 47, No. 11., pp. 747-757. | #[[http://udel.edu/~gshriver/pdf/Pimenteletal1997.pdf Pmentel et al., 1997]] Pimentel, D., Wilson, C., McCullum, C., Huang, R., Dwen, P., Flack, J. Tran, Q., Saltman, T., Cliff, T. (1997). Economic and environmental benefits of biodiversity. ''BioScience'', Vol. 47, No. 11., pp. 747-757. | ||
| - | + | ||
Revision as of 17:36, 26 September 2013
Characterization of recombinant effector proteins
Text. See proteinbiochemical methods for further informatons.
| Protein | BioBrick | RFC | Affinity tag | Size [kDa] | Disulphid bridges | comment | |
|---|---|---|---|---|---|---|---|
| Eryhtromycin esterase (EreB) | <partinfo>BBa_K1159000</partinfo> | RFC25 | c-term. Streptag II | ||||
| Laccase | <partinfo>BBa_K1159002</partinfo> | RFC25 | c-term. Streptag II | ||||
| Nano Luciferase | <partinfo>BBa_K1159001</partinfo> | RFC25 | c-term. Streptag II | ||||
| XylE | <partinfo>BBa_E0040</partinfo> | ||||||
| PP1 | <partinfo>BBa_K1159004</partinfo> | ||||||
| YFP_TEV_CFP | <partinfo>BBa_K1159112</partinfo> |
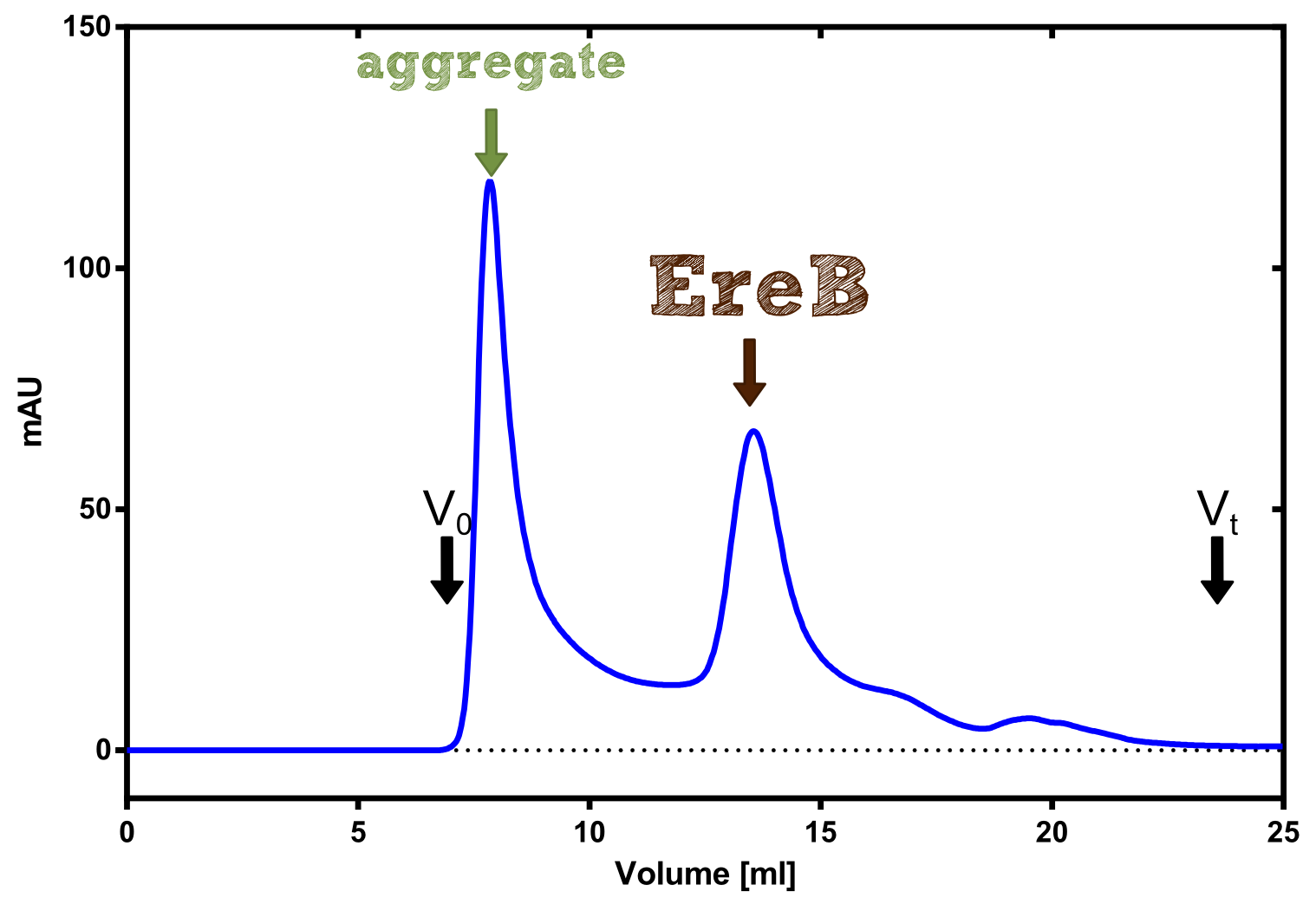
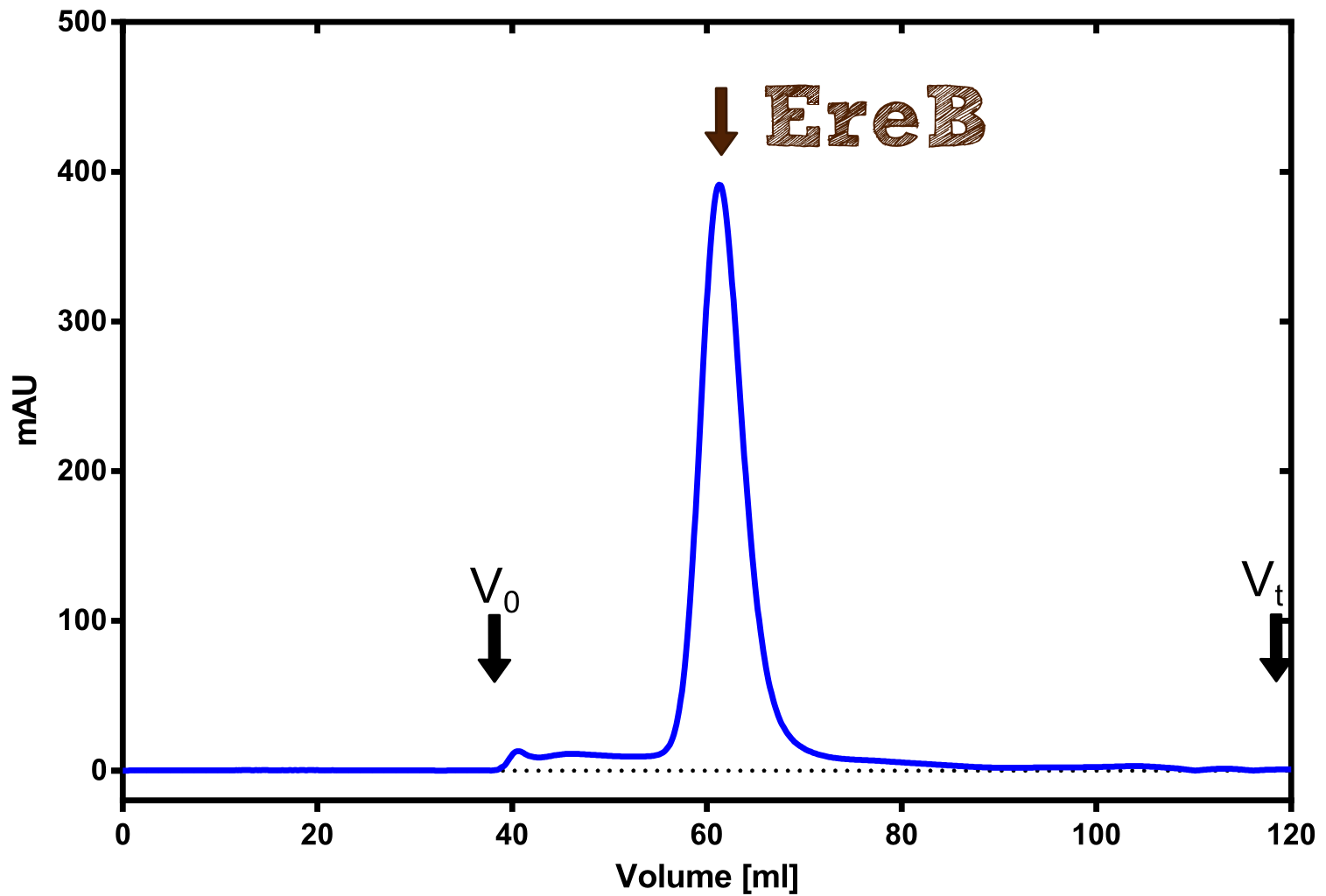
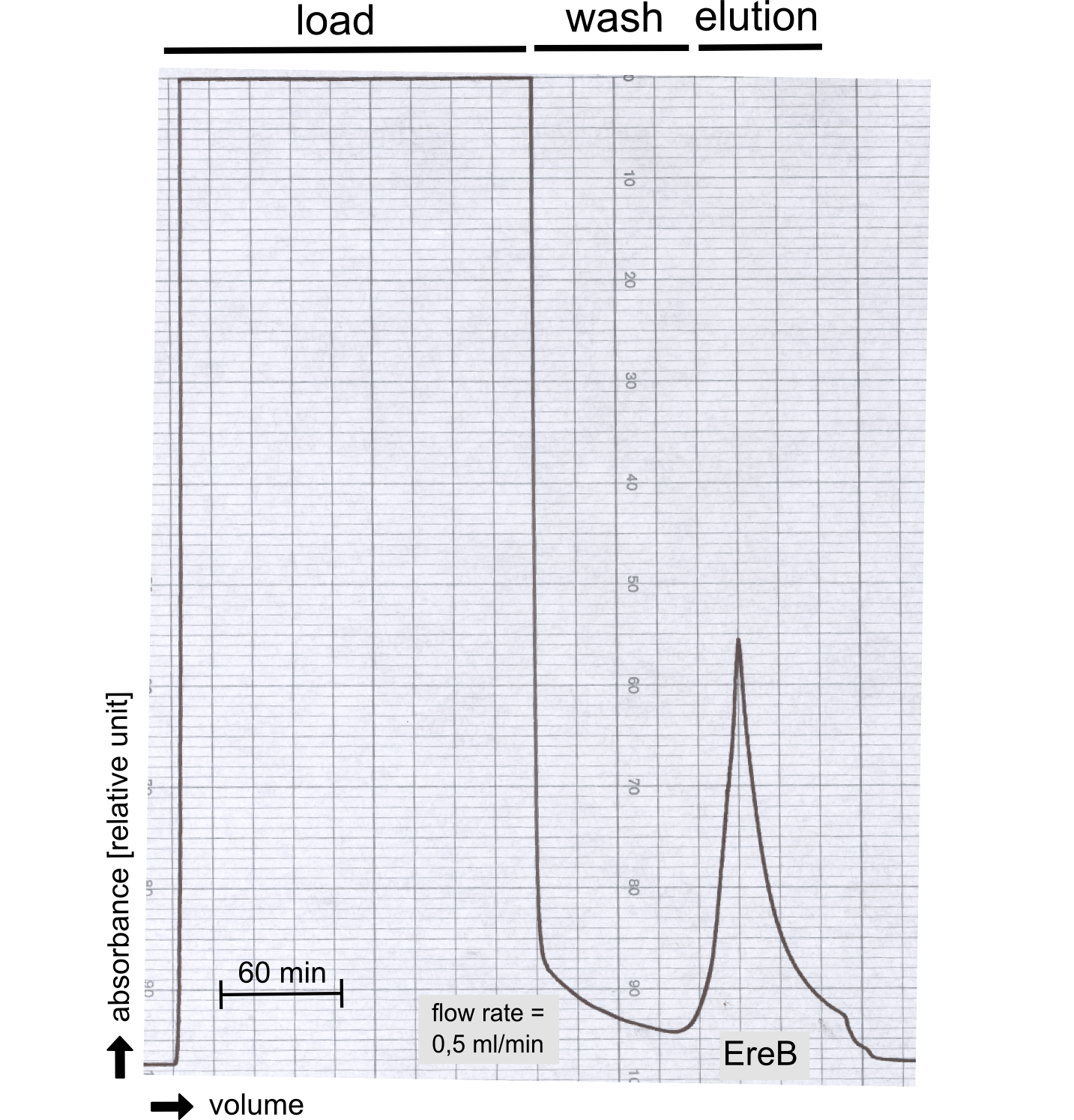
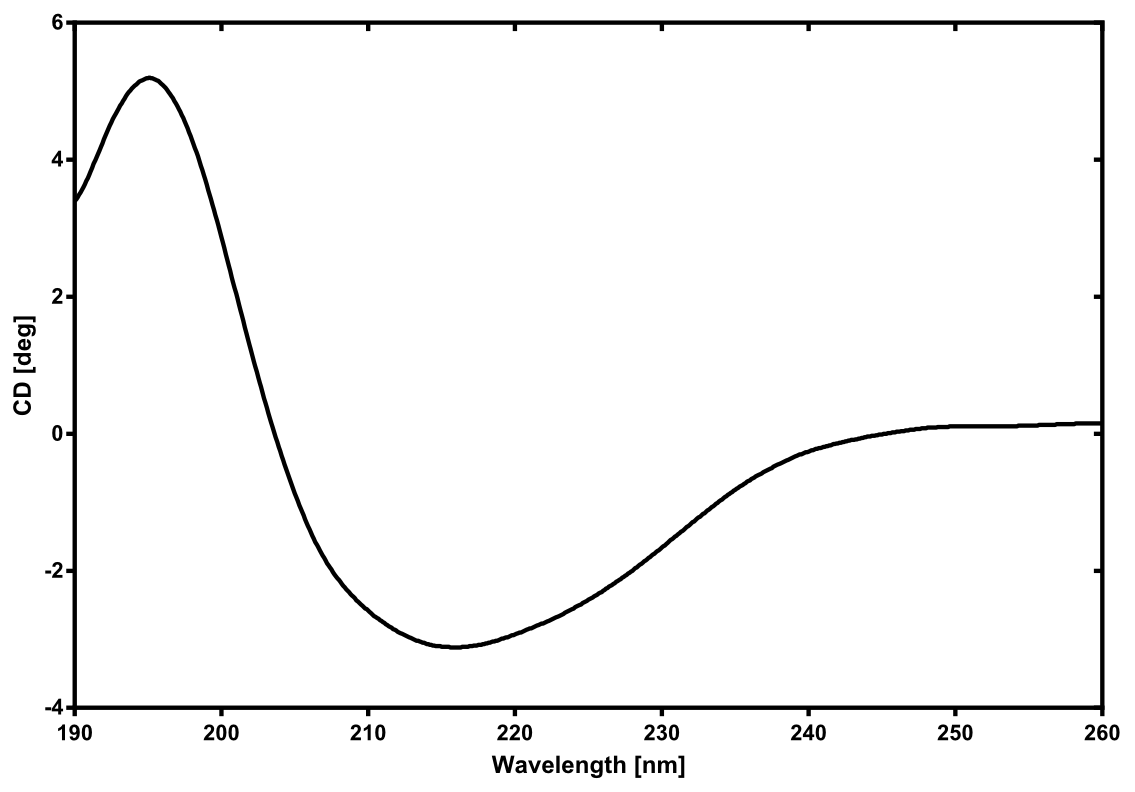
Eryhtromycin Esterase
[...] description [...] reaction [...] production
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCAGGTTCGAA ... GTTTATGAAACCGGT ORF from nucleotide position -8 to 1260 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Analytical präparation
[...] Characterization
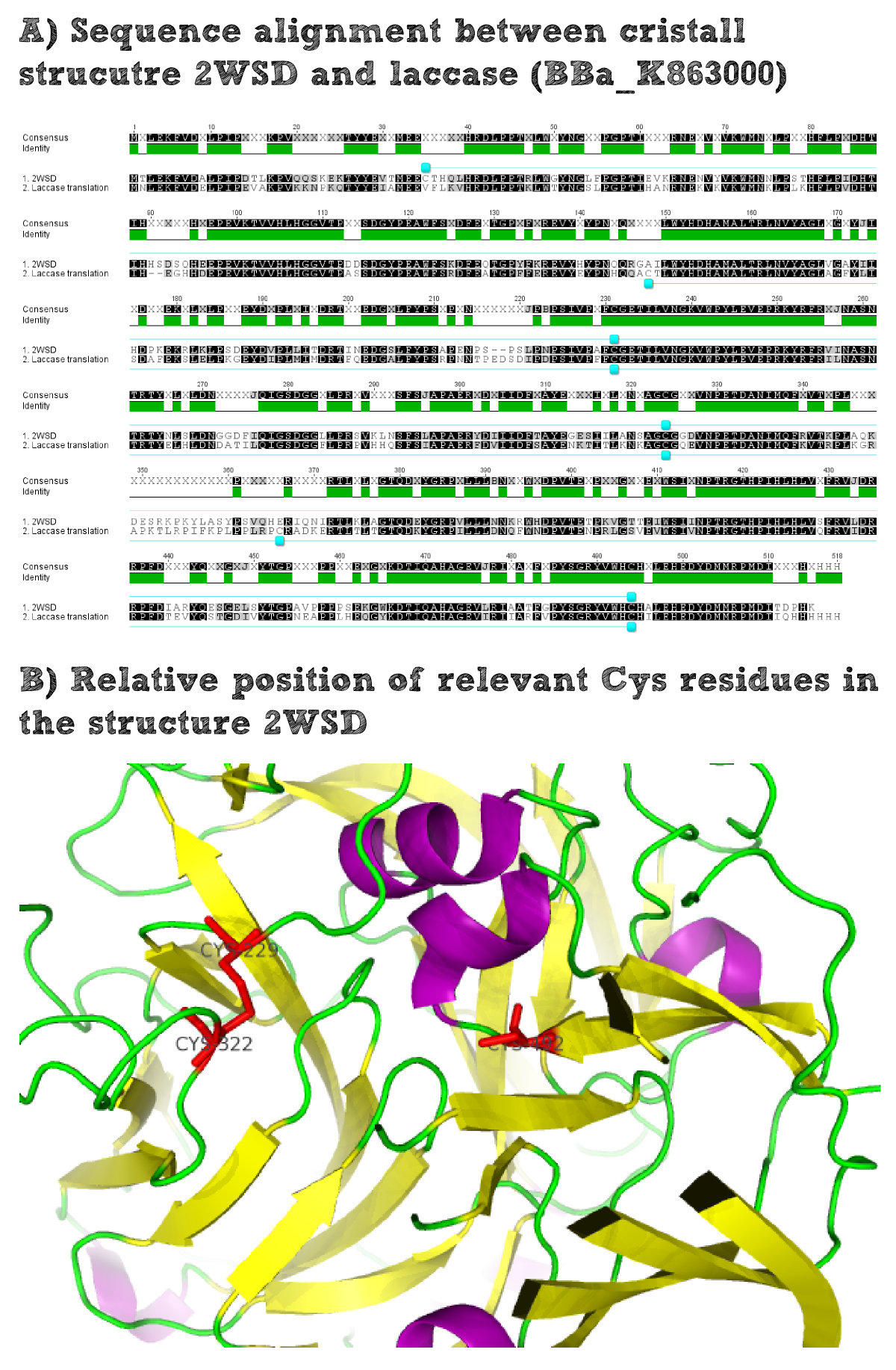
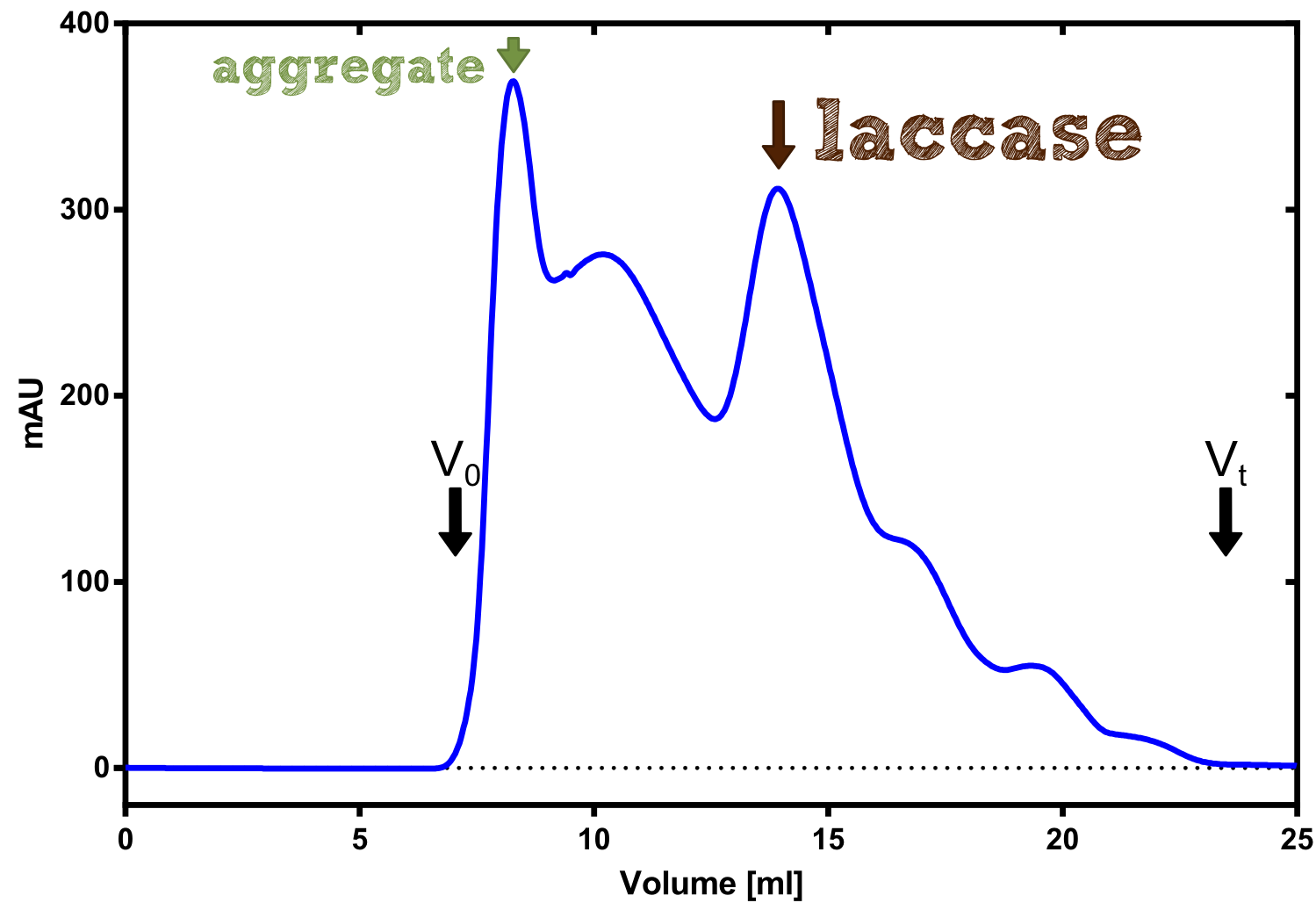
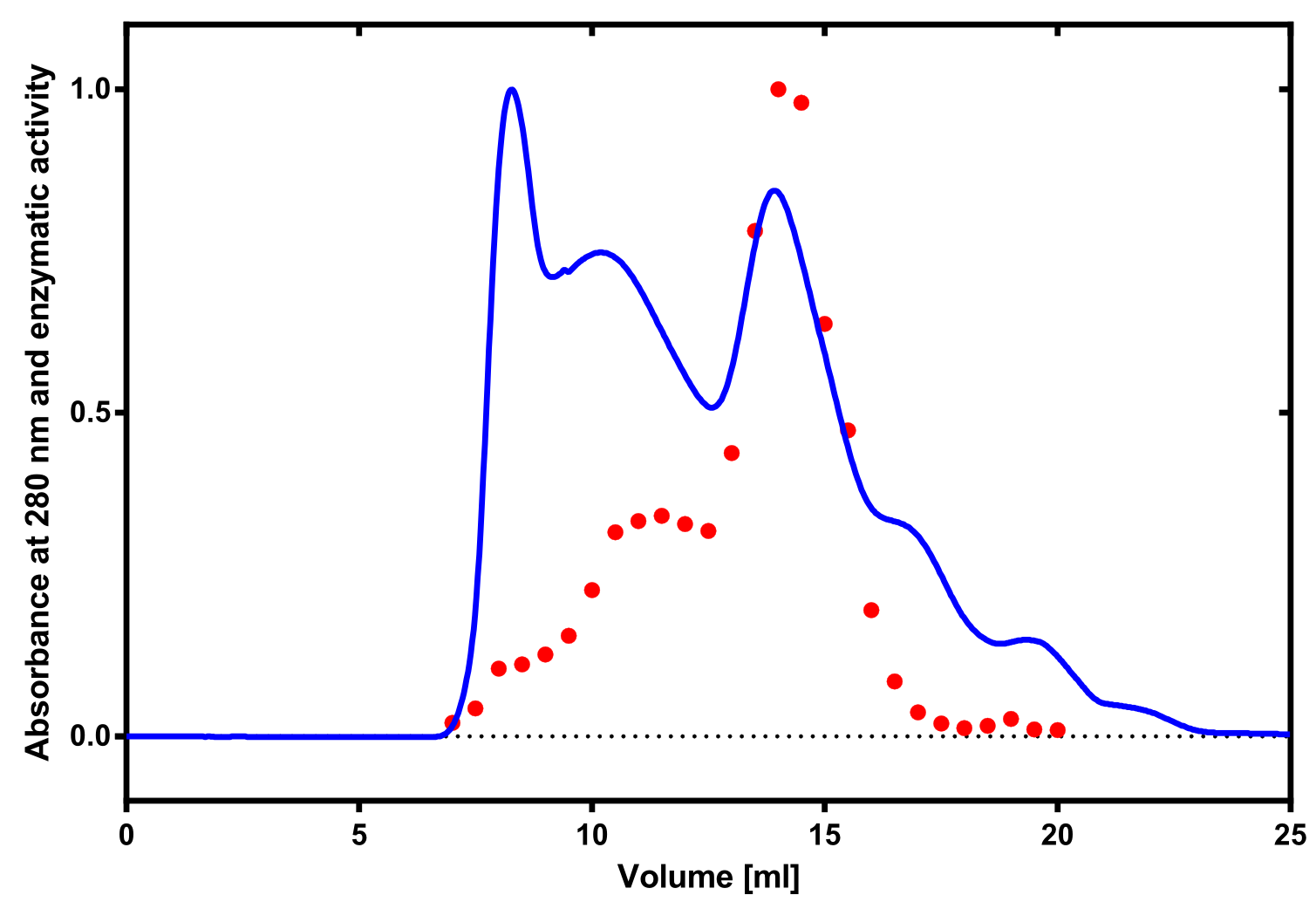
Laccase
[...] description [...] reaction [...] production
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCAACCTAGAA ... GATATCATCACCGGT ORF from nucleotide position -8 to 1530 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Laccase a is a secreted enzyme
--->
Analytical präparation
[...] Characterization
Aktivity determination using ABTS
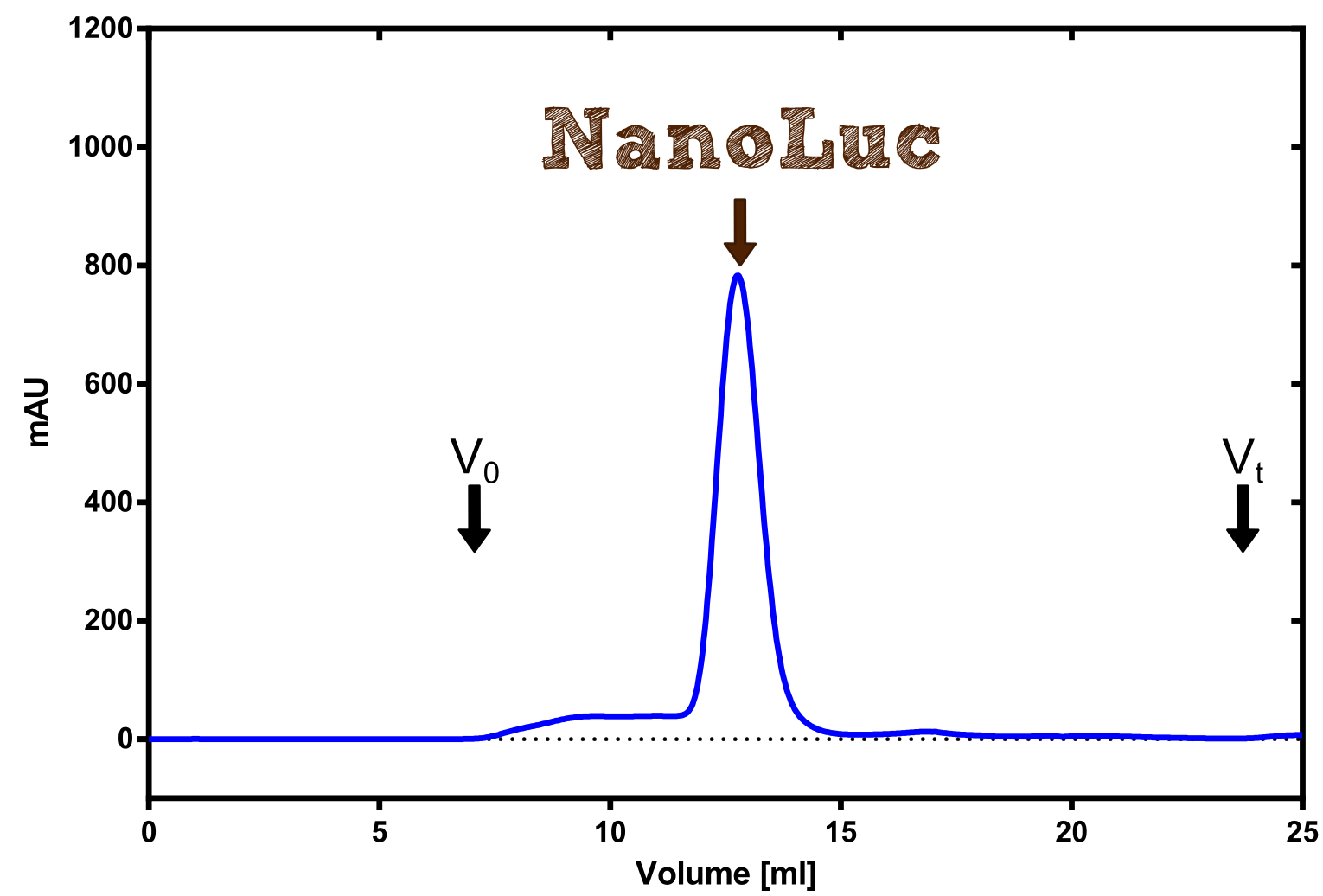
Nano Luciferase
[...] description [...] reaction [...] production
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGTCTTCACA ... ATTCTGGCGACCGGT ORF from nucleotide position -8 to 516 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Analytical präparation
[...] Characterization
Structure of the Nano Luciferase
--->
XylE
[...] description [...] reaction [...] production
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCAACAAAGGT ... GTGCTGACCACCGGT ORF from nucleotide position -8 to 924 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Analytical präparation
[...] Characterization
DDT-Dehydrochlorinase
[...] description [...] reaction [...] production
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 10: (underlined part encodes the protein) ATGGACTTT ... TTCCTGAGCTAGTAG ORF from nucleotide position 1 to 627 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Analytical präparation
[...] Characterization
PP1
SpyCatcher & SpyTag
[...] description [...] reaction [...] production
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGTTGATACC ... GCTCATATTACCGGT ORF from nucleotide position -8 to 345 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Analytical präparation
[...] Characterization
References:
http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984
- http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984 Edens, L., Bom, I., Ledeboer, A. M., Maat, J., Toonen, M. Y., Visser, C., and Verrips, C. T. (1984). Synthesis and processing of the plant protein thaumatin in yeast. Cell, 37(2):629–33.
- http://udel.edu/~gshriver/pdf/Pimenteletal1997.pdf Pmentel et al., 1997 Pimentel, D., Wilson, C., McCullum, C., Huang, R., Dwen, P., Flack, J. Tran, Q., Saltman, T., Cliff, T. (1997). Economic and environmental benefits of biodiversity. BioScience, Vol. 47, No. 11., pp. 747-757.
 "
"





AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351