Team:Bielefeld-Germany/Biosafety/Biosafety System L
From 2013.igem.org
| Line 9: | Line 9: | ||
<html> | <html> | ||
<style> | <style> | ||
| - | h1{padding:10px 0;padding-left: | + | h1{padding:10px 0;padding-left:175px; padding-right:170px; margin-top:70px; } |
.toc, #toc{width:255px;font-size:85%;} | .toc, #toc{width:255px;font-size:85%;} | ||
Revision as of 23:23, 29 September 2013
Biosafety System Lac of Growth
Overview
The tetracyclin repressor (TetR)/ operator (TetO) originally is used by E. coli to work against the antibiotic tetracycline but in many cases it is used for regulated expression for industrial processes. When there is no tetracycline available the TetR binds with high affinity the tetracycline operator. When tetracycline is available the TetR switches his conformation and so it comes to a dissolution of the TetR and the TetO. Because of this the polymerase isn’t enhanced anymore and is able to express the genes which lies behind the TetO. In our system the TetR is under the control of a rhamnose promotor (rha-promotor) which only works in the presence of rhamnose. When the bacteria would break out of the MFC there wouldn’t be enough rhamnose in the environment to activate the promotor in a way that enough TetR would be produced to block the polymerase by binding at the TetO. Therefore the polymerase binds to the promotor of TetO and it comes to the expression of RNase Ba and the degradation of the DNA.
Genetic Approach
lacI
The lac repressor/operator system uses E.coli to regulate the production of enzymes and because of this E. coli also regulate its metabolic stress. Enzymes only are produced when they are required. LacI is a repressor which is able to inhibit the lac operon in the absence of lactose by binding to the DNA at the lac operator site called lacO. Because of this repressor DNA polymerase is inhibited so it can’t read the sequences behind the operator lacZ (β-galactosidase), lacY (lactose permease) and lacA (thiogalactosidase transacetylase) which are responsible for transporting and metabolism of lactose in E. coli can’t be transcribed. The structure of the lac operon is shown in the image below.
Rhamnose
When L-rhamnose is in the milieu where E. coli is located it can be taken up by the RhaT transport system which converts it to L-rhamnulose by the isomerase RhaA. It continues by phosphorylating by the kinase RhaB in rhamnulose-1-phosphate. This is hydrolyzed by the aldolase RhaD into dihydroxyacetone phosphate and lactate aldehyde. Dihydroxyacetone is metabolized in glycolysis, and lactate aldehyde aerobe to lactate. If there are anaerobe conditions lactate aldehyde is reduced to L-1,2,-propandiol. The gene RhaBAD functions as an operon and is transcribed by RhaPBAD. Two activators, RhaR and RhaS, have to be expressed to regulate the system. This expression of these activators is in opposite direction than the expression of rhaBAD. When L-rhamnose is available RhaR binds to RhaPRS and activates the production of RhaR and RhaS. RhaS binds with L-rhamnose as an effector to RhaPBAD and RhaPT promoter and activates the transcription of the structural genes.
Alanine Racemase
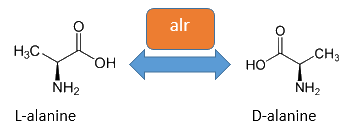
The alanine-racemase alr (EC 5.1.1.1) from the gram-negative enteric bacteria Escherichia coli is a racemase, which catalyses the reversible reaction from L-alanine into the enantiomer D-alanine. For this reaction the cofactor pyridoxal-5'-phosphate (PLP) is typically needed. The constitutive alanine-racemase (alr) is naturally responsible for the accumulation of D-Alanin, which is an essential component of the bacterial cell wall, because it is used for the crosslinkage of the peptidoglykan. The use of D-Alanin instead of a typically L-amino acids prevents the cleavage by peptdidases, but a lack of D-Alanin leeds to a bacteristatic characteristic. So in the absence of D‑Alanine dividing cells will lyse rapidly. So if the expression of the Alanin-Racemase is repressed and there is no D-Alanine-Supplementation in the media, the cells would not increase.
Barrnaaaaaase
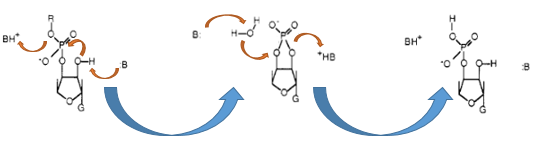
The Barnase (EC 3.1.27) is an 12 kDa extracellular microbial ribonuclease, which is naturally found in the gram-positive soilbacteria Bacillus amyloliquefaciens and consist a single chain of 110 amino acids. The Barnase (RNase Ba) catalyses the cleavage of single stranded RNA, where the hydrolysis of the dinucleotides has the highest affinity to the structure GpN. In the first step of the RNA-degradation a cyclic intermediate is formed by transesterification and afterwards this intermediate is hydrolysed yielding in a 3'-nucleotide (see graphic below).
In Bacillus amyloliquefaciens the activity ot the Barnase (RNase Ba) is inhibited intracelluar by the Inhibitor called barstar. Barstar consists only about 89 amnio acids and binds with a high affinity to the toxic Barnase. This prevents the cleavage of the intracellular RNA in the host organismen. Therefore the Barnase acts naturally only outside the cell and is translocated under natural conditions. For the Biosafety-System we tried to modified this aspect by cloning only the sequence responsbile for the cleavage of the RNA, but not the part of the native Barnase, which is essential for the extracellular transport. As shown in the graphic below, the transcription of the DNA, which encodes the Barnase produces a 474 nt RNA. The translation of the RNA of this Ribonuclease starts about 25 nucleotids downstream from the transcription start and can be divided into two parts. The first part (colored in orange) is translated into a signal peptide at the N-Terminus of the Barnase. This part is resopnsbile for the extracellular translocation of the RNase Ba, while the peptid sequence for the active Barnase starts 142 nucleotides downstream from the transcription start (colored in red). For the Biosafety-System we used only the coding sequence of the Barnase itself to prevent the extracellular translocation of the toxic gene product. This leds to a rapid cell death if the expression of the Barnase isn't repressed by the Repressor of the Biosafety-System.
Results
References
Agnes Ullmann (2001): Escherichia coli Lactose Operon. In: Encyclopedia of Life Sciences
Stumpp et al.: Ein neues, L-Rhamnose-induzierbares Expressionssystem für Escherichia coli, In: Biospektrum 6. Jahrgang S. 33
Carsten Voss, Dennis Lindau, and Erwin Flaschel, Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use, Biotechnology Progress, 22, 2006 p. 737-44.
Danuta E. Mossakowska, Kerstin Nyberg, and Alan R. Fersht, Kinetic Characterization of the Recombinant Ribonuclease from Bacillus amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis, Biochemistry, 28, 1989, p. 3843 – 3850.
C. J. Paddon, N. Vasantha, and R. W. Hartley, Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase, Journal of Bacteriology, 171, 1989, p. 1185 – 1187.
 "
"