Team:Bielefeld-Germany/Biosafety/Biosafety Strain
From 2013.igem.org
| Line 150: | Line 150: | ||
<p align="justify"> | <p align="justify"> | ||
| - | To verify the | + | To verify the deletion of the repressor araC in ''E. coli'' we designed some primer, which lay outside the deletion region so that a successfull deletion can be seen by an xx bp amplyfied DNA-fragmente, while the wildtype shows typically a DNA-fragement with a length of xx bp. The primers for the control are listed in the box below.</p> |
<br> | <br> | ||
Revision as of 21:42, 1 October 2013
Safety Strain
Overview
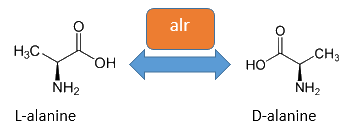
The basic for our Safety-System is an auxotropic Safety-Strain. In our Safety-Strain the constitutive Alanine-Racemase (alr) and the catabolic Alanine-Racemase (dadX) is deleted. As the Alanin-Racemase catalyses the reversible isomerization from L-alanine into the enantiomer D-alanine our safety-strain is not any more able to synthesis the essential amino acid D-alanine. In the gram-negative bacteria E. coli D-alanie is an essential molecul for the cross-linkage of the peptidoglycanlayer, so that a lack of D-alanine inhibits cell division and leads to cell lysis in growing bacteria. This is comparable to bacteristatic characteristic known from beta-lactam-antbiotics.
An important parameter, when expressing toxic gene products like the Rnase Ba, is the maintaince of the plasmidstability. So the strict dependance on D-alanine or complementation by the Alanin-Racemase via plasmid is used in our Safety-System to increase the stability of the plasmid, because in this case, not only the Barnase would imply cell death, but also the loss of the Alanine-Racemase (alr) from the plasmid.
Another deletion concerns the araC gene, the repressor of the arabinose promoter pBAD. As the araC protein functions not only as a repressor but also as an activator a strain with this mutation can be used to amplify a plasmid with a toxic gene product under the control of this promoter without causing cell death. This huge advantage is only possible, because the araC protein, as activator, is absolutely necessary for the initiation of transcription at this promoter. Besides the deletion of the araC gene decreases the amount of repressors when using the Biosafety-System so that the very tight repression of the pBAD promoter is a little bit relaxed and the expression of the toxic gene can so be fine tuned by investigation of different strains.
Theory
The cell wall is essential for every living bacteria, because it divides the Cytoplasm from the environment. In fact the life of the bacteria is only possible because this separation. So in one hand the cell wall confer stability and structure and on the other hand its possible to build up the osmotic pressure and to regulate the transport of molecules across the cell wall.
The composition of the cell wall differs between the bacteria, so that the bacteria in general are divided into gram-negative Bacteria, as example Escherichia coli and gram-positive Bacteria for example Bacillus subtillis. Gram-negative bacteria therefore characterized by an inner plasma membrane, a thin peptidoglycanlayer a periplasmatic space an the outer membrane. While gram-positive bacteria in contrast poses only one membrane but a thicker peptidoglycanlayer.
Therefore it becomes clear that the peptidoglycanlayer is an interesting approach to control the bacterial growth. Peptidoglycan itself is a polymer consisting linear chain of polysaccharides and short peptides. The polysaccharides component consists of alternating residues of beta-(1,4) linked N-acetylglucosamine and N-acetylmuramic acid and they are cross-linked in E. coli by a tetra-peptide of L-Alanine, D-glutamic acid, meso-diaminopimelic acid and final D-alanine. The cross-linkage is thereby realized by a transpeptid-linkage of meso-diaminopimelic acid and D-alanine.
This shows that D-Alanin is an essential component of the bacterial peptidoglycanlayer. In conclusion a lack of D-Alanin would have a bacteriostatic characteristic, comparable to beta-lactam-antibiotic, because it inhibits the building of the peptdiglycan-linkage. Therefore the bacteria without D-Alanin could not divide, because they will lyse otherwise.
The accumulation of D-alanine in E. coli can be catalyzed by an Alanine-Racemase (EC 5.1.1.1). This enzyme catalyses the reversible reaction from L-alanine into the enantiomer D-alanine. For this reaction the cofactor pyridoxal-5'-phosphate (PLP) is typically needed. In E. coli exists thereby two Alanine-Racemases. One Alanine-Racemase (alr) is constitutive expressed and therefore normally responsible for the accumulation of D-Alanine, while the Alanin-Racemase (dadX) is under control of the dad-Operon.
The deletion of the constitutive Alanin-Racemase (alr) and the catabolic Alanine-Racemase (dadX) will lead to a strict dependence of D-Alanine, so that the bacteria with this mutations only able to grow on media with D-alanine supplemented or by complementation of the Alanin-Racemase on a separate plasmid. As shown in the figure below the D-Alanin-Auxotrophic mutant shows a strict dependence on D-Alanine.</p>
Genetic Approach
For the construction of our Biosafety-System-Strain we deleted both Alanine-Racemases via homologous recombination with the pKO4 (a derivation from the pKO3 containing the coding sequence of the lacZ-alpha Fragment) to establish an D-alanine-auxotrophic mutant. For the deletion of the Alanine-Racemase we thereby used the following primers. The primers alr_Ec_d1 and alr_Ec_d2 will amplify an about 600 bp long homologous fragment upstream of the coding sequence of the Alanine-Racemase, while the primers alr_Ec_d3 and alr_Ec_d4 will perfom an about 600 bp long homologous fragment downstream of the Alanine-Racemase. This to deletion-sides can be ligated by Gibson-Assembly or alternative classically by gene SOEing (Gene splicing overlap Extension). The deletions-sides of the Alanine-Racemase were cloned into to vector by using the bold primer-overhang (see box below).
By following the protocol for the double-cross over using the heat-sensitive RepA and the sacB-Gene from the vector [http://arep.med.harvard.edu/labgc/pko3.html pKO3], the Alanine-Racemase could be successfully deleted.
Primer for the complete Deletion of the constitutive Alanine-Racemase (alr) and the catabolic Alanine-Racemase (dadX) in E. coli
Primer: alr_Ec_d1 (39 bp)
5'-TAGCTCACTCATTAGGCACCCAGCTCGATGACGAAGACT-3'
Primer: alr_Ec_d2 (20 bp)
5'-GCCGCTTGCATTTGTGTTCC-3'
Primer: alr_Ec_d3 (40 bp)
5'-GGAACACAAATGCAAGCGGCTTGATTGTCTGTGCCGGATG-3'
Primer: alr_Ec_d4 (40 bp)
5'-GCTTTCTACGTGTTCCGCTTCCGGGAAGTAGCGTTTCAGG-3'
Primer: dadX_Ec_d1 (39 bp)
5'-TAGCTCACTCATTAGGCACCTGAAGTGTGGCGATGAAGT-3'
Primer:dadX_Ec_d2 (20 bp)
5'-GGGTCATCTCGTTTCCTTAG-3'
Primer: dadX_Ec_d3 (40 bp)
5'-CTAAGGAAACGAGATGACCCACTTGTTGTAAGCCGGATCG-3'
Primer: dadX_Ec_d4 (40 bp)
5'-GCTTTCTACGTGTTCCGCTTCGAAGCCAGCGCCAAATATG-3'
To verify the Deletion of the Alanine-Racemase in E. coli we designed some primer, which lay outside the deletion region so that a successfull deletion can be seen by an xx bp amplyfied DNA-fragmente, while the wildtype shows typically a DNA-fragement with a length of xx bp. The primers for the control are listed in the box below.
Primer for the control of the successful Deletion of the constitutive Alanine-Racemase (alr) and the catabolic Alanine-Racemase (dadX) in E. coli
Primer: alr_Ec_control1 (20 bp)
5'-GCTGGAGATGCCATCAGAAC-3'
Primer: alr_Ec_control2 (20 bp)
5'-CCGGCGAATATTGCTACGTG-3'
Primer: dadX_Ec_control1 (20 bp)
5'-GCTTTAATACGCCCGTTGAC-3'
Primer: dadX_Ec_control2 (20 bp)
5'-CTGGATCAACGCTTCTTTGG-3'
For the Deletion of the repressor araC we therefore used the same method as for the deletion auf die Alanin-Racemases in E. coli. The primers araC_d1 and araC_d2 will amplifiy analogous an about 600 bp long homologous fragment upstream of the coding sequence of the coding sequence for araC, while the primers araC_d3 and araC_d4 will perfom an about 600 bp long homologous fragment downstream of the araC gene. This two deletion-sides can also be ligated by Gibson-Assembly or gene SOEing (Gene splicin overlap Extension) and then cloned into to vector by using the bold primer-overhang (see box below). By following the protocol for the double-cross over using the heat-sensitive RepA and the sacB-gene from the vector pKO3, the Repressor araC could be successfully deleted.
Primer for the complete Deletion of the repressor araC for the arabinose promoter pBAD in E. coli
Primer: araC_d1 (40 bp)
5'-TAGCTCACTCATTAGGCACCCCGGCAGGAATATCGATCGC-3'
Primer: araC_d2 (40 bp)
5'-CTTCTCTGAATGGTGGGAGTGTCATAATTGGTAACGAATC-3'
Primer: araC_d3 (40 bp)
5'-GATTCGTTACCAATTATGACACTCCCACCATTCAGAGAAG-3'
Primer: araC_d4 (40 bp)
5'-GCTTTCTACGTGTTCCGCTTAACGCCAATCCCGACCACAG-3'
To verify the deletion of the repressor araC in E. coli we designed some primer, which lay outside the deletion region so that a successfull deletion can be seen by an xx bp amplyfied DNA-fragmente, while the wildtype shows typically a DNA-fragement with a length of xx bp. The primers for the control are listed in the box below.
Primer for the control of the successful Deletion of repressor araC in E. coli
Primer: araC_control1 (20 bp)
5'-CTTTACCGCTGCGCCATAAC-3'
Primer: araC_control2 (20 bp)
5'-AACCGCAGTGTGGTCTTTCC-3'
Results
References
- Link, A.J., Phillips, D. and Church, G.M. (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC179534/pdf/1796228.pdf|Journal of Bacteriology 179: 6228-6237.]
- Autoren (Jahr) Titel [Link|Paper Ausgabe: Seiten].
 "
"