Team:Bielefeld-Germany/Project/Porins
From 2013.igem.org
| Line 65: | Line 65: | ||
==Overview - Porin OprF== | ==Overview - Porin OprF== | ||
| + | |||
[[Image:Bielefeld-germany-project-overview-porine.png|300px|left|thumb|'''Figure 1:''' Schematic of the enhancement mechanism of electron shuttle-mediated electron transfer between bacteria and the anode of MFCs by the synthetic porin OprF.]] | [[Image:Bielefeld-germany-project-overview-porine.png|300px|left|thumb|'''Figure 1:''' Schematic of the enhancement mechanism of electron shuttle-mediated electron transfer between bacteria and the anode of MFCs by the synthetic porin OprF.]] | ||
| + | |||
<p align="justify"> | <p align="justify"> | ||
The Microbial Fuel Cell (MFC) can be a future environmental friendly biotechnology for production of electrical energy. A major limiting factor is the low bacterial membrane permeability, limiting transport of electron shuttles through the membrane that restricts the electron shuttle-mediated extracellular electron transfer (EET) from bacteria to electrodes. This results in a reduced electrical power output of the MFC. Therefore, we heterologously expressed the porin protein OprF from ''Pseudomonas fluorescens'' into ''Escherichia coli''. This leads to dramatically increased membrane permeability and a much higher current output in comparison to its parental strain (''E. coli'' KRX) caused by improved electron shuttle-mediated extracellular electron transfer. The heterologous expression of outer membrane porin OprF from ''Pseudomonas fluorescens'' in ''Escherichia coli'' is a great genetic strategy to improve electricity generation by microorganisms. | The Microbial Fuel Cell (MFC) can be a future environmental friendly biotechnology for production of electrical energy. A major limiting factor is the low bacterial membrane permeability, limiting transport of electron shuttles through the membrane that restricts the electron shuttle-mediated extracellular electron transfer (EET) from bacteria to electrodes. This results in a reduced electrical power output of the MFC. Therefore, we heterologously expressed the porin protein OprF from ''Pseudomonas fluorescens'' into ''Escherichia coli''. This leads to dramatically increased membrane permeability and a much higher current output in comparison to its parental strain (''E. coli'' KRX) caused by improved electron shuttle-mediated extracellular electron transfer. The heterologous expression of outer membrane porin OprF from ''Pseudomonas fluorescens'' in ''Escherichia coli'' is a great genetic strategy to improve electricity generation by microorganisms. | ||
</p> | </p> | ||
| - | |||
<br><br> | <br><br> | ||
| Line 77: | Line 78: | ||
==Theory== | ==Theory== | ||
| + | |||
| + | |||
*<p align="justify">The efficiency of extracellular electron transfer is a major limiting factor for electricity power output of MFCs. The electron shuttle-mediated EET is the most common EET pathway for microorganisms in a Microbial Fuel Cell such as ''Escherichia coli'' ([[Team:Bielefeld-Germany/Project/Porins#References|Logan, 2009]]).</p> | *<p align="justify">The efficiency of extracellular electron transfer is a major limiting factor for electricity power output of MFCs. The electron shuttle-mediated EET is the most common EET pathway for microorganisms in a Microbial Fuel Cell such as ''Escherichia coli'' ([[Team:Bielefeld-Germany/Project/Porins#References|Logan, 2009]]).</p> | ||
*<p align="justify">The cell membrane is a natural protective layer enabling proper physiology of bacteria, but it is also a barrier for substrates’ exchange, because an efficient electron shuttle-mediated EET requires diffusion of shuttle molecules across cell membrane (Figure 1). However, the bacterial outer membrane is a low permeable barrier for the transport of electron shuttles across the cell membrane, which really limits the efficiency of electron transport and is responsible for the low power output of MFCs up to now. One strategy to improve the electron shuttle mediated EET is to enhance the permeability of cell membrane. Evolutionary strategies for example continuous cell stress are very time-consuming and unpredictable. The chemical treatment with permeabilizers can perforate the outer membrane but has negative impact on the viability and metabolism of the cells. ([[Team:Bielefeld-Germany/Project/Porins#References|Liu ''et al''., 2012]])</p> | *<p align="justify">The cell membrane is a natural protective layer enabling proper physiology of bacteria, but it is also a barrier for substrates’ exchange, because an efficient electron shuttle-mediated EET requires diffusion of shuttle molecules across cell membrane (Figure 1). However, the bacterial outer membrane is a low permeable barrier for the transport of electron shuttles across the cell membrane, which really limits the efficiency of electron transport and is responsible for the low power output of MFCs up to now. One strategy to improve the electron shuttle mediated EET is to enhance the permeability of cell membrane. Evolutionary strategies for example continuous cell stress are very time-consuming and unpredictable. The chemical treatment with permeabilizers can perforate the outer membrane but has negative impact on the viability and metabolism of the cells. ([[Team:Bielefeld-Germany/Project/Porins#References|Liu ''et al''., 2012]])</p> | ||
| Line 95: | Line 98: | ||
==Results== | ==Results== | ||
===Overview=== | ===Overview=== | ||
| - | |||
| + | |||
| + | *<p align="justify">Upon the expression of the outer membrane porin protein OprF, the morphology and physicochemical characteristics of the ''E. coli'' surface were measured. [[Team:Bielefeld-Germany/Labjournal/Molecular#Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) | SDS-PAGE]] combined with [[Team:Bielefeld-Germany/Labjournal/Molecular#MALDI-TOF|MALDI-TOF MS/MS]], different membrane permeability assays ([[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN]] and [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay | ONPG]]), a [[Team:Bielefeld-Germany/Labjournal/Molecular#Hexadecan Assay|hydrophobicity assay]] and [[Team:Bielefeld-Germany/Labjournal/Molecular#Preparing samples for Atomic Force Microscopy (AFM) analysis of the OM of cells|Atomic Force Microscopy]] (AFM) characterize OprF BioBrick <bbpart>BBa_K1172501</bbpart>.</p> | ||
===SDS-PAGE and MALDI-TOF=== | ===SDS-PAGE and MALDI-TOF=== | ||
| + | |||
| + | |||
*<p align="justify">OprF could been detected with [[Team:Bielefeld-Germany/Labjournal/Molecular#Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)|SDS-PAGE]]. Protein isolation of the outer membrane porin OprF by Release of periplasmic protein fraction from ''E. coli'' by [[Team:Bielefeld-Germany/Labjournal/Molecular#Cold osmotic shock|cold osmotic shock]] using [[Team:Bielefeld-Germany/Labjournal/Molecular#Cell Fractioning Buffers | Cell fractionating buffer 2.3]] was successful.</p> | *<p align="justify">OprF could been detected with [[Team:Bielefeld-Germany/Labjournal/Molecular#Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)|SDS-PAGE]]. Protein isolation of the outer membrane porin OprF by Release of periplasmic protein fraction from ''E. coli'' by [[Team:Bielefeld-Germany/Labjournal/Molecular#Cold osmotic shock|cold osmotic shock]] using [[Team:Bielefeld-Germany/Labjournal/Molecular#Cell Fractioning Buffers | Cell fractionating buffer 2.3]] was successful.</p> | ||
[[Image:IGEM_Bielefeld_SDS_PAGE_OprF3.jpg|300px|thumb|left|<p align="justify"> '''Figure 2: [[Team:Bielefeld-Germany/Labjournal/Molecular#Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)|SDS-PAGE]] with [http://www.thermoscientific.com/ecomm/servlet/productsdetail_11152___13576050_-1 Prestained Protein Ladder from Thermo Scientific] as marker. Comparison of protein expression between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart> and <bbpart>BBa_K1172507</bbpart> after [[Team:Bielefeld-Germany/Labjournal/Molecular#Cold osmotic shock | periplasmic protein fractioning]] with [[Team:Bielefeld-Germany/Labjournal/Molecular#Cell Fractioning Buffers | Cell fractionating buffer 2.3]]. '''</p>]] | [[Image:IGEM_Bielefeld_SDS_PAGE_OprF3.jpg|300px|thumb|left|<p align="justify"> '''Figure 2: [[Team:Bielefeld-Germany/Labjournal/Molecular#Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)|SDS-PAGE]] with [http://www.thermoscientific.com/ecomm/servlet/productsdetail_11152___13576050_-1 Prestained Protein Ladder from Thermo Scientific] as marker. Comparison of protein expression between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart> and <bbpart>BBa_K1172507</bbpart> after [[Team:Bielefeld-Germany/Labjournal/Molecular#Cold osmotic shock | periplasmic protein fractioning]] with [[Team:Bielefeld-Germany/Labjournal/Molecular#Cell Fractioning Buffers | Cell fractionating buffer 2.3]]. '''</p>]] | ||
| + | |||
| + | |||
*<p align="justify">The [[Team:Bielefeld-Germany/Labjournal/Molecular#Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)|SDS-PAGE]] shows a significantly higher protein concentration for ''E.coli'' with OprF and T7 promoter (<bbpart>BBa_K1172502</bbpart>). It seems to be that the higher membrane permeability (shown with [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN]] and [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG]] uptake assay) allows a better release of membrane proteins by 0.2 % SDS. Nevertheless, we can see a strong overexpression band at the expected OprF size of about 36 kDa for <bbpart>BBa_K1172502</bbpart>, which is equated with a strong expression and overproduction of OprF.</p> | *<p align="justify">The [[Team:Bielefeld-Germany/Labjournal/Molecular#Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)|SDS-PAGE]] shows a significantly higher protein concentration for ''E.coli'' with OprF and T7 promoter (<bbpart>BBa_K1172502</bbpart>). It seems to be that the higher membrane permeability (shown with [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN]] and [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG]] uptake assay) allows a better release of membrane proteins by 0.2 % SDS. Nevertheless, we can see a strong overexpression band at the expected OprF size of about 36 kDa for <bbpart>BBa_K1172502</bbpart>, which is equated with a strong expression and overproduction of OprF.</p> | ||
*<p align="justify">Furthermore we were able to identify the overexpressed outer membrane porin (Figure. 2) with [[Team:Bielefeld-Germany/Labjournal/Molecular#MALDI-TOF|MALDI-TOF MS/MS]].</p> | *<p align="justify">Furthermore we were able to identify the overexpressed outer membrane porin (Figure. 2) with [[Team:Bielefeld-Germany/Labjournal/Molecular#MALDI-TOF|MALDI-TOF MS/MS]].</p> | ||
| Line 110: | Line 118: | ||
===Hydrophobicity (Hexadecane) - Assay=== | ===Hydrophobicity (Hexadecane) - Assay=== | ||
| + | |||
| + | |||
*<p align="justify">The heterologous expression of the outer membrane porin OprF will enhance the hydrophobicity of cell membrane. The outward-facing side groups on each of the β-strands of the OprF monomer are hydrophobic. Therefore a positive expression should be visible by an increase in hydrophobicity.</p> | *<p align="justify">The heterologous expression of the outer membrane porin OprF will enhance the hydrophobicity of cell membrane. The outward-facing side groups on each of the β-strands of the OprF monomer are hydrophobic. Therefore a positive expression should be visible by an increase in hydrophobicity.</p> | ||
*<p align="justify">An increasing hydrophobicity of cell membrane changes the physicochemical properties of the cell. This could effect for example the cell-electrode interaction. Therefore, we investigated the cellular surface characteristic by comparing ''Escherichia coli'' KRX wildtyp with ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart> (Table 2 and Figure 3).</p> | *<p align="justify">An increasing hydrophobicity of cell membrane changes the physicochemical properties of the cell. This could effect for example the cell-electrode interaction. Therefore, we investigated the cellular surface characteristic by comparing ''Escherichia coli'' KRX wildtyp with ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart> (Table 2 and Figure 3).</p> | ||
| - | *<p align="justify">The OprF strain shows an increasing affinity to hexadecane with increasing promotor strength in comparison to the Wildtyp. OprF with T7 promotor (<bbpart>BBa_K1172502</bbpart>) shows the maximal hydrophobicity which was three times higher than affinity to hexadecane of the Wildtyp. | + | *<p align="justify">The OprF strain shows an increasing affinity to hexadecane with increasing promotor strength in comparison to the Wildtyp. OprF with T7 promotor (<bbpart>BBa_K1172502</bbpart>) shows the maximal hydrophobicity which was three times higher than affinity to hexadecane of the Wildtyp.</p> |
| + | |||
| + | |||
[[Image:IGEM_Bielefeld_Table5_Hexadecan.jpg|300px|thumb|left|<p align="justify"> '''Table 2: Results of the [[Team:Bielefeld-Germany/Labjournal/Molecular#Hexadecan Assay | Hexadecane-Hydrophobicity-Assay]]. Comparison of protein Hydrophobicity between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Affinity to hexadecane (Hydrophobicity) with standard deviation and enhancement in comparison to the wild-type is shown. '''</p>]] | [[Image:IGEM_Bielefeld_Table5_Hexadecan.jpg|300px|thumb|left|<p align="justify"> '''Table 2: Results of the [[Team:Bielefeld-Germany/Labjournal/Molecular#Hexadecan Assay | Hexadecane-Hydrophobicity-Assay]]. Comparison of protein Hydrophobicity between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Affinity to hexadecane (Hydrophobicity) with standard deviation and enhancement in comparison to the wild-type is shown. '''</p>]] | ||
| - | [[Image:IGEM_Bielefeld_Figure7_Results_Hexadecane.jpg|300px|thumb|center|<p align="justify"> '''Figure 3: Results of the [[Team:Bielefeld-Germany/Labjournal/Molecular#Hexadecan Assay | Hexadecane-Hydrophobicity-Assay]]. Comparison of protein Hydrophobicity between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Affinity to hexadecane (Hydrophobicity) with standard deviation and enhancement in comparison to the wild-type is shown. '''</p>]] | + | [[Image:IGEM_Bielefeld_Figure7_Results_Hexadecane.jpg|300px|thumb|center|<p align="justify"> '''Figure 3: Results of the [[Team:Bielefeld-Germany/Labjournal/Molecular#Hexadecan Assay | Hexadecane-Hydrophobicity-Assay]]. Comparison of protein Hydrophobicity between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Affinity to hexadecane (Hydrophobicity) with standard deviation and enhancement in comparison to the wild-type is shown. '''</p>]] |
| Line 124: | Line 136: | ||
===Membrane permeabilty assays=== | ===Membrane permeabilty assays=== | ||
====NPN uptake assay==== | ====NPN uptake assay==== | ||
| + | |||
| + | |||
*<p align="justify">Besides testing the outer membrane hydrophobicity for physicochemical characterization of the ''E. coli'' surface, we measured the membrane permeability by [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay | NPN uptake assay]] for outer membrane morphology characterization (Helander and Mattila-Sandholm, 2000).</p> | *<p align="justify">Besides testing the outer membrane hydrophobicity for physicochemical characterization of the ''E. coli'' surface, we measured the membrane permeability by [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay | NPN uptake assay]] for outer membrane morphology characterization (Helander and Mattila-Sandholm, 2000).</p> | ||
*<p align="justify">NPN is a very suitable chemical for measuring the membrane permeability of cells. An increasing NPN fluorescence intensity indicates an enhanced NPN uptaking by the outer membrane and enhanced membrane permeability (Loh et al., 1984).</p> | *<p align="justify">NPN is a very suitable chemical for measuring the membrane permeability of cells. An increasing NPN fluorescence intensity indicates an enhanced NPN uptaking by the outer membrane and enhanced membrane permeability (Loh et al., 1984).</p> | ||
*<p align="justify">Figure 8 shows a higher fluorescence emission and therefore higher membrane permeability with increasing promotor strength for OprF strains in comparison to ''Escherichia coli'' KRX wildtyp. | *<p align="justify">Figure 8 shows a higher fluorescence emission and therefore higher membrane permeability with increasing promotor strength for OprF strains in comparison to ''Escherichia coli'' KRX wildtyp. | ||
| + | |||
| + | |||
[[Image:IGEM_Bielefeld_Figure8_Results_NPN2.jpg|450px|thumb|left|<p align="justify"> '''Figure 4: Results of the [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay | NPN-uptake-assay]]. Comparison of fluorescence emission between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Excitation at 355 nm with fluorescence emission scan from 320 up to 390 nm wavelength and with standard deviation. '''</p>]]</p> | [[Image:IGEM_Bielefeld_Figure8_Results_NPN2.jpg|450px|thumb|left|<p align="justify"> '''Figure 4: Results of the [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay | NPN-uptake-assay]]. Comparison of fluorescence emission between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Excitation at 355 nm with fluorescence emission scan from 320 up to 390 nm wavelength and with standard deviation. '''</p>]]</p> | ||
| + | |||
| + | |||
*<p align="justify">''Escherichia coli'' with heterologous expression of the outer membrane porin OprF shows more efficient NPN uptaking than the ''Escherichia coli'' KRX Wildtyp, suggesting an increasing membrane permeability with increasing promotor strength. ''E. coli'' with T7 promotor (<bbpart>BBa_K1172502</bbpart>) shows the maximal membrane permeability with 100% enhanced permeability in comparison to ''Escherichia coli'' KRX Wildtyp. The weak Anderson promoters seem unsuitable for heterologous expression.</p> | *<p align="justify">''Escherichia coli'' with heterologous expression of the outer membrane porin OprF shows more efficient NPN uptaking than the ''Escherichia coli'' KRX Wildtyp, suggesting an increasing membrane permeability with increasing promotor strength. ''E. coli'' with T7 promotor (<bbpart>BBa_K1172502</bbpart>) shows the maximal membrane permeability with 100% enhanced permeability in comparison to ''Escherichia coli'' KRX Wildtyp. The weak Anderson promoters seem unsuitable for heterologous expression.</p> | ||
====ONPG assay==== | ====ONPG assay==== | ||
| + | |||
| + | |||
*<p align="justify">Another way of characterizing the outer cell membrane can be achieved by [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]]. [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]] measures the membrane permeability by whole-cell lactase enzyme activity (Zhou et al., 2010).</p> | *<p align="justify">Another way of characterizing the outer cell membrane can be achieved by [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]]. [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]] measures the membrane permeability by whole-cell lactase enzyme activity (Zhou et al., 2010).</p> | ||
*<p align="justify">Electron shuttle-mediated electron transfer (EET) determines the transport efficiency of electron shuttles across the cell membrane. With [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN uptake assay]] we were able to quantify the transport efficiency of chemical molecules across the cell membrane. [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]] (Whole-cell β-lactase enzyme activity assay) can show furthermore the diffusion of ONPG hydrolytic product out of the cell membrane, which quantifies not only uptaking of chemical molecules, but also the hydrolytic product secretion. With [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]] we are able to observe diffusion processes in and out of the cell membrane.</p> | *<p align="justify">Electron shuttle-mediated electron transfer (EET) determines the transport efficiency of electron shuttles across the cell membrane. With [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN uptake assay]] we were able to quantify the transport efficiency of chemical molecules across the cell membrane. [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]] (Whole-cell β-lactase enzyme activity assay) can show furthermore the diffusion of ONPG hydrolytic product out of the cell membrane, which quantifies not only uptaking of chemical molecules, but also the hydrolytic product secretion. With [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]] we are able to observe diffusion processes in and out of the cell membrane.</p> | ||
| - | *<p align="justify">Due the fact that heterologous expression of OprF in ''Escherichia coli'' has no impact on the expression of β-lactase, we can assume that the β-lactase activity of ''Escherichia coli'' KRX Wildtyp and ''Escherichia coli'' KRX with OprF expression plasmid is identical. All in all, the [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]] provides significantly more information than the [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN uptake assay]]. | + | *<p align="justify">Due the fact that heterologous expression of OprF in ''Escherichia coli'' has no impact on the expression of β-lactase, we can assume that the β-lactase activity of ''Escherichia coli'' KRX Wildtyp and ''Escherichia coli'' KRX with OprF expression plasmid is identical. All in all, the [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]] provides significantly more information than the [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN uptake assay]].</p> |
| - | [[Image:IGEM_Bielefeld_Figure9_Results_ONPG2.jpg|450px|thumb|left|<p align="justify"> '''Figure 5: Results of the [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG-uptake-assay]]. Comparison of ONPG hydrolysis between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Absorbance at 405 nm wavelength with standard deviation is shown. '''</p>]] | + | |
| + | |||
| + | [[Image:IGEM_Bielefeld_Figure9_Results_ONPG2.jpg|450px|thumb|left|<p align="justify"> '''Figure 5: Results of the [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG-uptake-assay]]. Comparison of ONPG hydrolysis between ''Escherichia coli'' KRX wild type and ''Escherichia coli'' KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Absorbance at 405 nm wavelength with standard deviation is shown. '''</p>]] | ||
| + | |||
| + | |||
*<p align="justify">The ONPG hydrolysis rate by β-lactase is much higher for ''Escherichia coli'' KRX with OprF plasmids in contrast to ''Escherichia coli'' KRX Wildtyp. Whereas we couldn’t observe any hydrolysis activity for Wildtyp. ''Escherichia coli'' KRX with OprF plasmids shows a much faster ONPG hydrolysis rate with increasing promotor strength and a maximal rate for'' Escherichia coli'' KRX with T7 promotor (<bbpart>BBa_K1172502</bbpart>), which is 30 times higher than Wildtyp hydrolysis rate.</p> | *<p align="justify">The ONPG hydrolysis rate by β-lactase is much higher for ''Escherichia coli'' KRX with OprF plasmids in contrast to ''Escherichia coli'' KRX Wildtyp. Whereas we couldn’t observe any hydrolysis activity for Wildtyp. ''Escherichia coli'' KRX with OprF plasmids shows a much faster ONPG hydrolysis rate with increasing promotor strength and a maximal rate for'' Escherichia coli'' KRX with T7 promotor (<bbpart>BBa_K1172502</bbpart>), which is 30 times higher than Wildtyp hydrolysis rate.</p> | ||
| + | |||
*<p align="justify">In summary we can say that the heterologous expression of OprF from ''Pseudomonas fluorescens'' in ''Escherichia coli'' significantly improves the membrane permeability. [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN]] and [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG]] assay show correlating results. ''Escherichia coli'' outer membrane permeability is enhanced with increasing promotor strength for OprF expression. The Wildtyp shows only a weak uptake rate of chemical molecules (NPN) but no product secretion as quantified with [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]]. Therefore, Wildtyp is not suitable for a usage in the MFC. Whereas membrane optimized ''Escherichia coli'' with OprF shows great diffusion processes in and out of the cellular membrane, indicating a great optimization of electron shuttle-mediated electron transfer (EET) to the anode and increasing current production.</p> | *<p align="justify">In summary we can say that the heterologous expression of OprF from ''Pseudomonas fluorescens'' in ''Escherichia coli'' significantly improves the membrane permeability. [[Team:Bielefeld-Germany/Labjournal/Molecular#NPN membrane permeability assay|NPN]] and [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG]] assay show correlating results. ''Escherichia coli'' outer membrane permeability is enhanced with increasing promotor strength for OprF expression. The Wildtyp shows only a weak uptake rate of chemical molecules (NPN) but no product secretion as quantified with [[Team:Bielefeld-Germany/Labjournal/Molecular#ONPG membrane permeability assay |ONPG assay]]. Therefore, Wildtyp is not suitable for a usage in the MFC. Whereas membrane optimized ''Escherichia coli'' with OprF shows great diffusion processes in and out of the cellular membrane, indicating a great optimization of electron shuttle-mediated electron transfer (EET) to the anode and increasing current production.</p> | ||
| Line 142: | Line 167: | ||
===Atomic Force Microscopy (AFM)=== | ===Atomic Force Microscopy (AFM)=== | ||
| + | |||
| + | |||
*<p align="justify">In addition to morphology and physicochemical characterization of the ''Escherichia coli'' outer membrane, we wanted to visualize the surface. The technique of choice for this is Atomic Force Microscopy (AFM). [[Team:Bielefeld-Germany/Labjournal/Molecular#Preparing samples for Atomic Force Microscopy (AFM) analysis of the OM of cells|After cell preparation]] we were able to get AFM pictures of ''E. coli'' surface with the help of the working group of [http://www.physik.uni-bielefeld.de/biophysik Prof. Dr. Dario Anselmetti], with special help from [http://www.physik.uni-bielefeld.de/biophysik/mitarbeiter/walhorn.html Dr. Volker Walhorn]. Thank you very much for your help!</p> | *<p align="justify">In addition to morphology and physicochemical characterization of the ''Escherichia coli'' outer membrane, we wanted to visualize the surface. The technique of choice for this is Atomic Force Microscopy (AFM). [[Team:Bielefeld-Germany/Labjournal/Molecular#Preparing samples for Atomic Force Microscopy (AFM) analysis of the OM of cells|After cell preparation]] we were able to get AFM pictures of ''E. coli'' surface with the help of the working group of [http://www.physik.uni-bielefeld.de/biophysik Prof. Dr. Dario Anselmetti], with special help from [http://www.physik.uni-bielefeld.de/biophysik/mitarbeiter/walhorn.html Dr. Volker Walhorn]. Thank you very much for your help!</p> | ||
| Line 163: | Line 190: | ||
===Microbial Fuel Cell Measurement=== | ===Microbial Fuel Cell Measurement=== | ||
| + | |||
| + | |||
*<p align="justify">All results described by different evidence indicate an increase in energy production. All these assumptions were confirmed by cultivation of'' Escherichia coli'' KRX with OprF (<bbpart>BBa_K1172502</bbpart>) in contrast to ''Escherichia coli'' KRX Wildtyp in the Microbial Fuel Cell.</p> | *<p align="justify">All results described by different evidence indicate an increase in energy production. All these assumptions were confirmed by cultivation of'' Escherichia coli'' KRX with OprF (<bbpart>BBa_K1172502</bbpart>) in contrast to ''Escherichia coli'' KRX Wildtyp in the Microbial Fuel Cell.</p> | ||
*<p align="justify">According to our assumptions, the extracellular electron transfer mediated by electron shuttles is improved in the OprF strain resulting in an increased bioelectricity output. (Figure 10 and 11)</p> | *<p align="justify">According to our assumptions, the extracellular electron transfer mediated by electron shuttles is improved in the OprF strain resulting in an increased bioelectricity output. (Figure 10 and 11)</p> | ||
| Line 176: | Line 205: | ||
===Conclusion=== | ===Conclusion=== | ||
| + | |||
| + | |||
*<p align="justify">Cell membranes are protective layers which are crucial for physiology of the bacteria. In consideration of the Microbial Fuel Cell, membrane is also a barrier which decreases mediator and thus electron exchange between bacteria and anode. With heterologous expression of the outer membrane porin OprF from ''Pseudomonas fluorescens'' in ''Escherichia coli'' we are able to enhance membrane permeability. With this optimized cell membrane surface we generate an efficient electron shuttle-mediated EET with decreased limitation of EET by membrane barrier. In contrast to a perforation of the membrane with chemicals, cell viability is maintained with OprF expression (Liu et al., 2012). Besides improvement of EET, enhanced hydrophobicity shows optimized cell adhesion to the anode for biofilm formation and direct electron transfer.</p> | *<p align="justify">Cell membranes are protective layers which are crucial for physiology of the bacteria. In consideration of the Microbial Fuel Cell, membrane is also a barrier which decreases mediator and thus electron exchange between bacteria and anode. With heterologous expression of the outer membrane porin OprF from ''Pseudomonas fluorescens'' in ''Escherichia coli'' we are able to enhance membrane permeability. With this optimized cell membrane surface we generate an efficient electron shuttle-mediated EET with decreased limitation of EET by membrane barrier. In contrast to a perforation of the membrane with chemicals, cell viability is maintained with OprF expression (Liu et al., 2012). Besides improvement of EET, enhanced hydrophobicity shows optimized cell adhesion to the anode for biofilm formation and direct electron transfer.</p> | ||
| + | |||
*<p align="justify">The heterologous expression of OprF is a great genetic strategy to optimize electron shuttle-mediated electron transfer as well as electricity generation in Microbial Fuel Cells. The most suitable and efficient OprF device for ''Escherichia coli'' is a combination with Rhamnose inducible T7 promotor (<bbpart>BBa_K1172502</bbpart>).</p> | *<p align="justify">The heterologous expression of OprF is a great genetic strategy to optimize electron shuttle-mediated electron transfer as well as electricity generation in Microbial Fuel Cells. The most suitable and efficient OprF device for ''Escherichia coli'' is a combination with Rhamnose inducible T7 promotor (<bbpart>BBa_K1172502</bbpart>).</p> | ||
| Line 182: | Line 214: | ||
==References== | ==References== | ||
| + | |||
*<p align="justify">Hancock REW, Brinkman FSL (2002) Function of ''Pseudomonas'' porins in uptake and efflux. [http://www.annualreviews.org/doi/abs/10.1146/annurev.micro.56.012302.160310| ''Annu Rev Microbiol'' (56):17–38].</p> | *<p align="justify">Hancock REW, Brinkman FSL (2002) Function of ''Pseudomonas'' porins in uptake and efflux. [http://www.annualreviews.org/doi/abs/10.1146/annurev.micro.56.012302.160310| ''Annu Rev Microbiol'' (56):17–38].</p> | ||
Revision as of 17:08, 2 October 2013
Porins
Overview - Porin OprF
The Microbial Fuel Cell (MFC) can be a future environmental friendly biotechnology for production of electrical energy. A major limiting factor is the low bacterial membrane permeability, limiting transport of electron shuttles through the membrane that restricts the electron shuttle-mediated extracellular electron transfer (EET) from bacteria to electrodes. This results in a reduced electrical power output of the MFC. Therefore, we heterologously expressed the porin protein OprF from Pseudomonas fluorescens into Escherichia coli. This leads to dramatically increased membrane permeability and a much higher current output in comparison to its parental strain (E. coli KRX) caused by improved electron shuttle-mediated extracellular electron transfer. The heterologous expression of outer membrane porin OprF from Pseudomonas fluorescens in Escherichia coli is a great genetic strategy to improve electricity generation by microorganisms.
Theory
The efficiency of extracellular electron transfer is a major limiting factor for electricity power output of MFCs. The electron shuttle-mediated EET is the most common EET pathway for microorganisms in a Microbial Fuel Cell such as Escherichia coli (Logan, 2009).
The cell membrane is a natural protective layer enabling proper physiology of bacteria, but it is also a barrier for substrates’ exchange, because an efficient electron shuttle-mediated EET requires diffusion of shuttle molecules across cell membrane (Figure 1). However, the bacterial outer membrane is a low permeable barrier for the transport of electron shuttles across the cell membrane, which really limits the efficiency of electron transport and is responsible for the low power output of MFCs up to now. One strategy to improve the electron shuttle mediated EET is to enhance the permeability of cell membrane. Evolutionary strategies for example continuous cell stress are very time-consuming and unpredictable. The chemical treatment with permeabilizers can perforate the outer membrane but has negative impact on the viability and metabolism of the cells. (Liu et al., 2012)
We thus hypothesize that genetic engineering of a highly permeable cell membrane would be a great idea. Therefore we introduce pore forming proteins in the outer membrane. So called porins are common bacterial outer membrane proteins, which can form water-filled channels across the membrane of gram-negative bacteria. They allow hydrophilic substances to diffuse across outer membrane (Hancock and Brinkman, 2002).
E. coli expresses different own porines, for example OmpF and OmpC. But these naturally occurring porines are only permeable for molecules smaller than 600 Da, which decreases the range of usable mediators and the mediator transport kinetic (Yong et al., 2013). Opposed to that, we thought about enhancing the amount of pores in the outer cell membrane with much larger porins than these of E. coli.
OprF is a major outer membrane protein in Pseudomonas species, which is a non-specific porin protein and adhesin. OprF presents one of the largest pore sizes on bacterial outer membrane that allows the diffusion of polysaccharides of 2000 up to 3000 Da, in contrast to the general porin channels of E. coli that are only permeable to sugars with sizes smaller than 600 Da. (Yong et al., 2013)
In due consideration of all facts, heterologous expression of the porin protein OprF from Pseudomonas fluorescens in E. coli will improve the EET between bacteria and electrodes by increase of membrane permeability. A heterologous expression of large porins improves besides mediator effectivity the spectrum of possible usable mediators. Thus environmentally friendly mediators such as NADH and riboflavin could be used significantly improved.
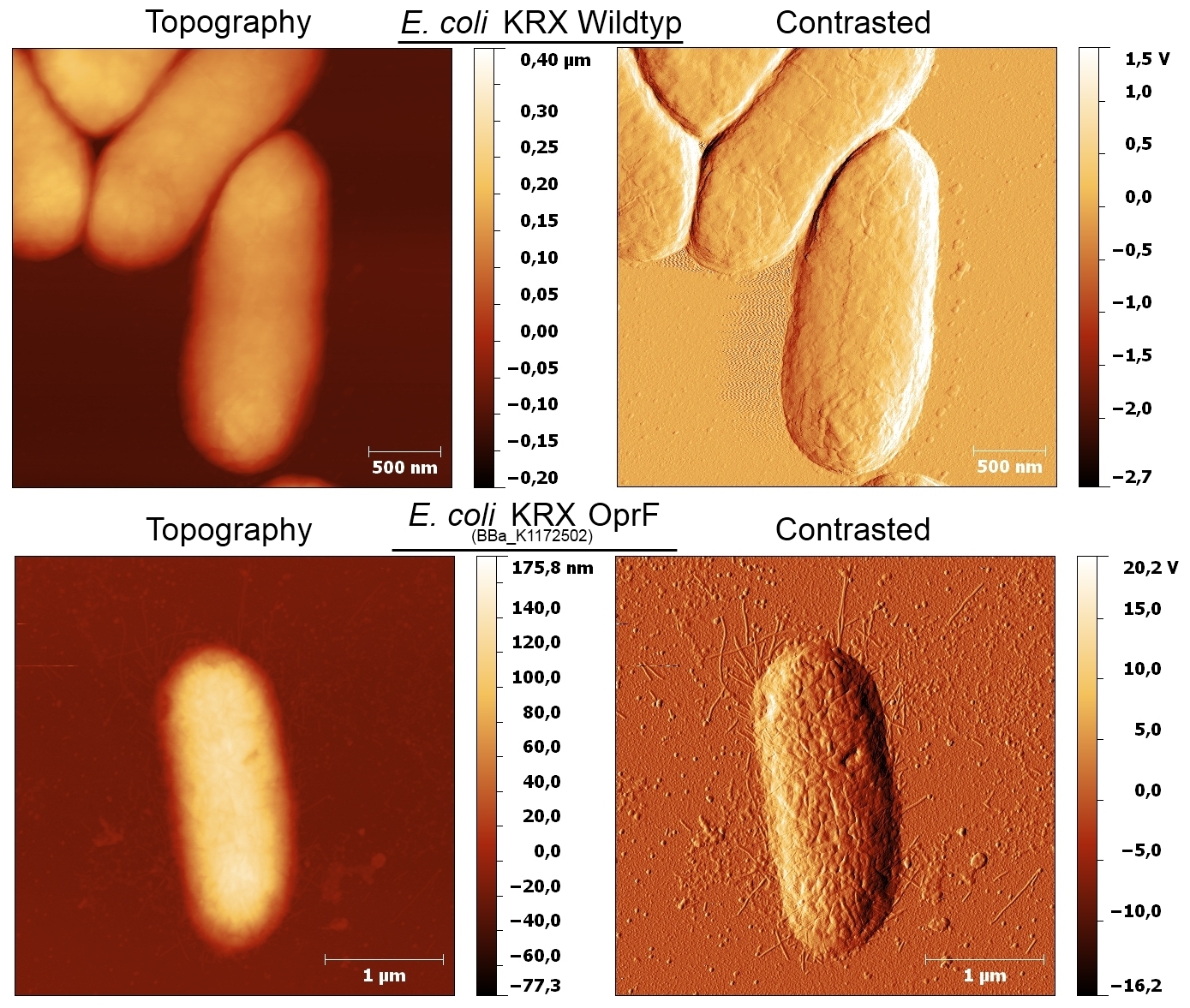
Genetic Approach
The OprF gene from Pseudomonas fluorescens was cloned and heterologously expressed in Escherichia coli KRX under the control of different promoters (Table 1).
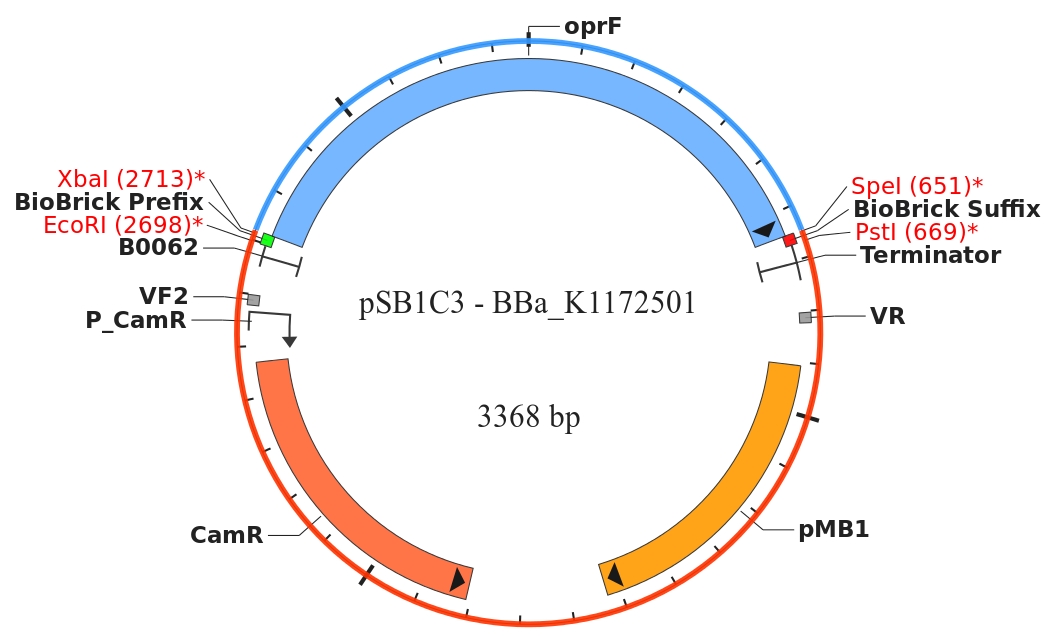
Figure 1: pSB1C3 – <bbpart>BBa_K1172501</bbpart> OprF BioBrick (1298 bp) was examined by restriction analysis and sequencing.
Results
Overview
Upon the expression of the outer membrane porin protein OprF, the morphology and physicochemical characteristics of the E. coli surface were measured. SDS-PAGE combined with MALDI-TOF MS/MS, different membrane permeability assays (NPN and ONPG), a hydrophobicity assay and Atomic Force Microscopy (AFM) characterize OprF BioBrick <bbpart>BBa_K1172501</bbpart>.
SDS-PAGE and MALDI-TOF
OprF could been detected with SDS-PAGE. Protein isolation of the outer membrane porin OprF by Release of periplasmic protein fraction from E. coli by cold osmotic shock using Cell fractionating buffer 2.3 was successful.
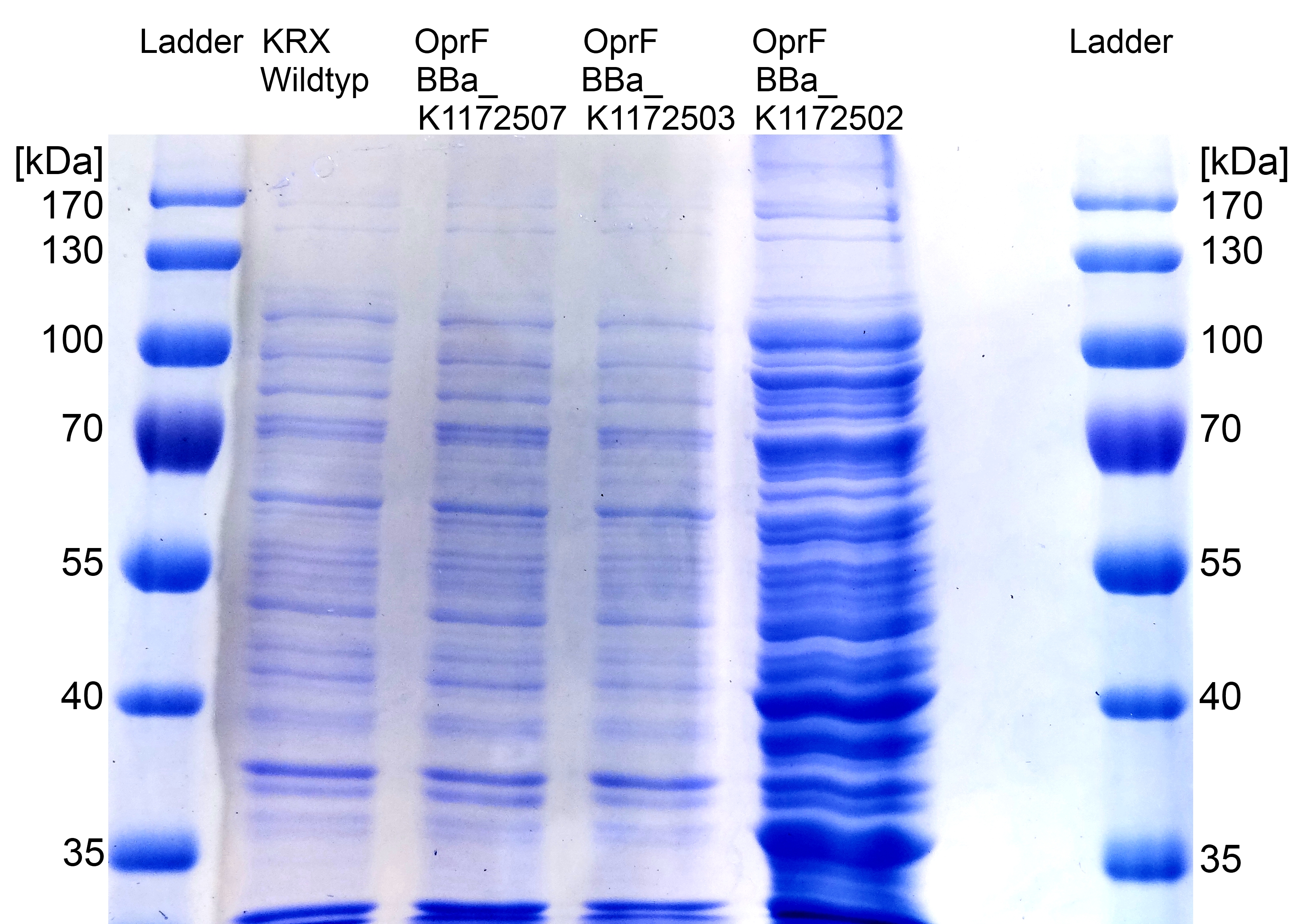
Figure 2: SDS-PAGE with [http://www.thermoscientific.com/ecomm/servlet/productsdetail_11152___13576050_-1 Prestained Protein Ladder from Thermo Scientific] as marker. Comparison of protein expression between Escherichia coli KRX wild type and Escherichia coli KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart> and <bbpart>BBa_K1172507</bbpart> after periplasmic protein fractioning with Cell fractionating buffer 2.3.
The SDS-PAGE shows a significantly higher protein concentration for E.coli with OprF and T7 promoter (<bbpart>BBa_K1172502</bbpart>). It seems to be that the higher membrane permeability (shown with NPN and ONPG uptake assay) allows a better release of membrane proteins by 0.2 % SDS. Nevertheless, we can see a strong overexpression band at the expected OprF size of about 36 kDa for <bbpart>BBa_K1172502</bbpart>, which is equated with a strong expression and overproduction of OprF.
Furthermore we were able to identify the overexpressed outer membrane porin (Figure. 2) with MALDI-TOF MS/MS.
Tryptic digest of the gel lane for analysis with MALDI-TOF could examine the outer membrane porin with a Mascot Score of 222 against bacteria database.
Hydrophobicity (Hexadecane) - Assay
The heterologous expression of the outer membrane porin OprF will enhance the hydrophobicity of cell membrane. The outward-facing side groups on each of the β-strands of the OprF monomer are hydrophobic. Therefore a positive expression should be visible by an increase in hydrophobicity.
An increasing hydrophobicity of cell membrane changes the physicochemical properties of the cell. This could effect for example the cell-electrode interaction. Therefore, we investigated the cellular surface characteristic by comparing Escherichia coli KRX wildtyp with Escherichia coli KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart> (Table 2 and Figure 3).
The OprF strain shows an increasing affinity to hexadecane with increasing promotor strength in comparison to the Wildtyp. OprF with T7 promotor (<bbpart>BBa_K1172502</bbpart>) shows the maximal hydrophobicity which was three times higher than affinity to hexadecane of the Wildtyp.
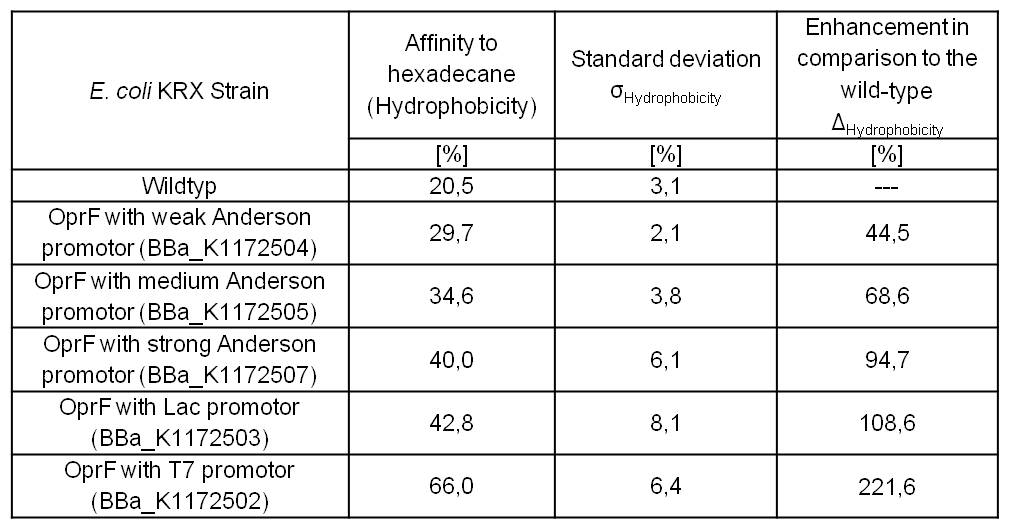
Table 2: Results of the Hexadecane-Hydrophobicity-Assay. Comparison of protein Hydrophobicity between Escherichia coli KRX wild type and Escherichia coli KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Affinity to hexadecane (Hydrophobicity) with standard deviation and enhancement in comparison to the wild-type is shown.
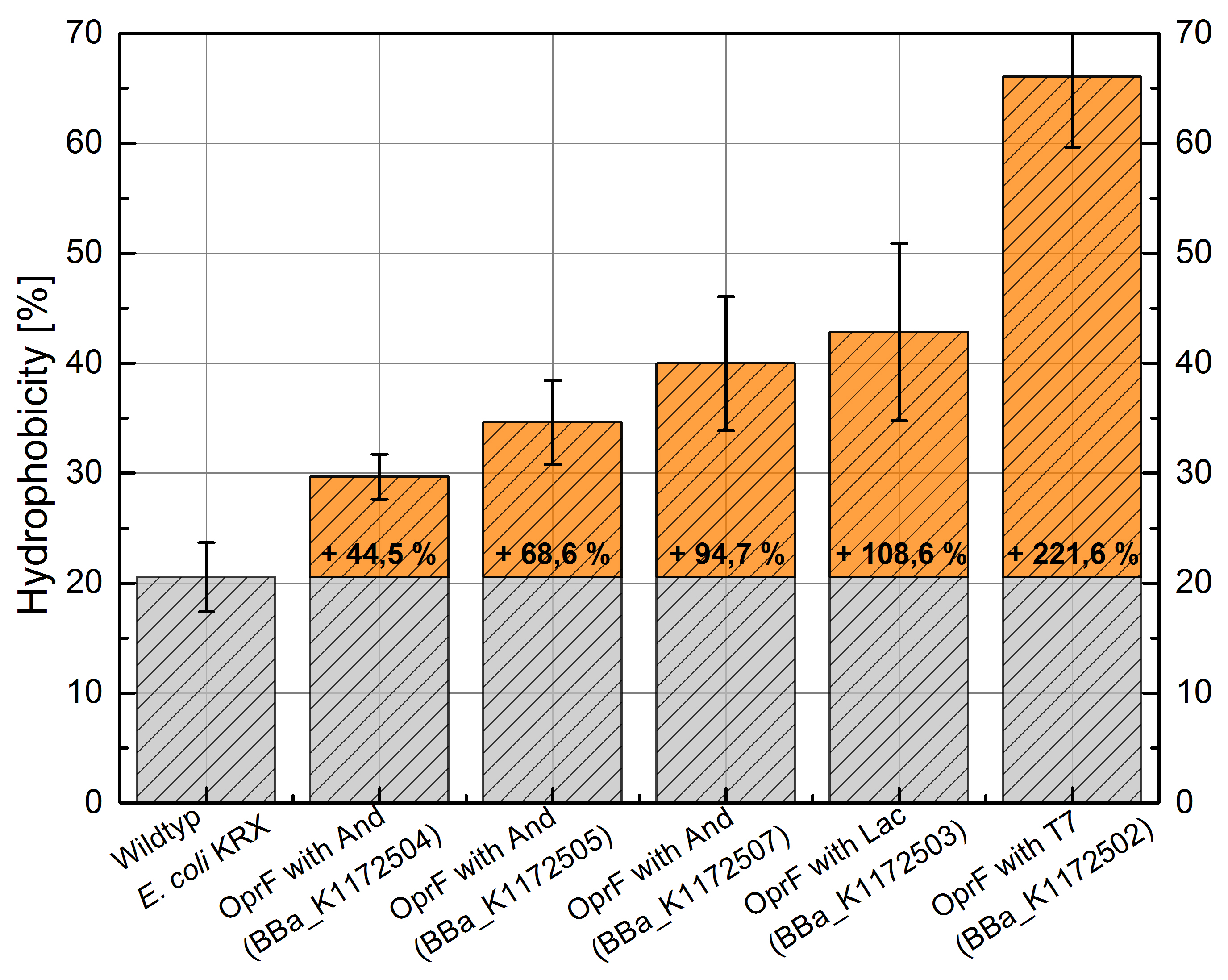
Figure 3: Results of the Hexadecane-Hydrophobicity-Assay. Comparison of protein Hydrophobicity between Escherichia coli KRX wild type and Escherichia coli KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Affinity to hexadecane (Hydrophobicity) with standard deviation and enhancement in comparison to the wild-type is shown.
The enhanced hydrophobicity of OprF-strains indicates a successful expression of the outer membrane porin in Escherichia coli. Such an increased hydrophobicity on the outer membrane caused by the expression of OprF will lead to an increase in the cellular adhesion to the surface of the carbon anode and an enhancement of direct electron transfer from Escherichia coli to the electrode.
Membrane permeabilty assays
NPN uptake assay
Besides testing the outer membrane hydrophobicity for physicochemical characterization of the E. coli surface, we measured the membrane permeability by NPN uptake assay for outer membrane morphology characterization (Helander and Mattila-Sandholm, 2000).
NPN is a very suitable chemical for measuring the membrane permeability of cells. An increasing NPN fluorescence intensity indicates an enhanced NPN uptaking by the outer membrane and enhanced membrane permeability (Loh et al., 1984).
Figure 8 shows a higher fluorescence emission and therefore higher membrane permeability with increasing promotor strength for OprF strains in comparison to Escherichia coli KRX wildtyp.
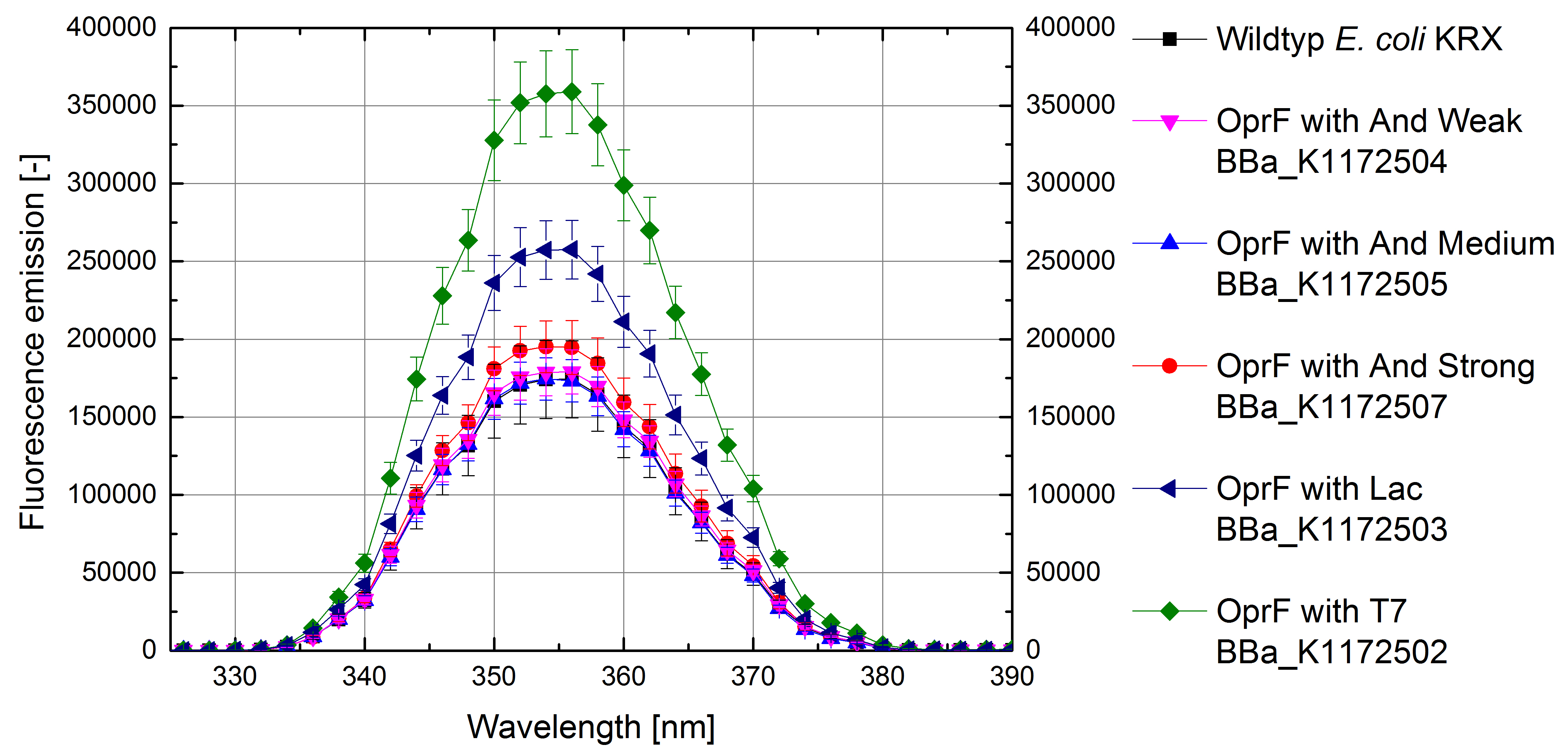
Escherichia coli with heterologous expression of the outer membrane porin OprF shows more efficient NPN uptaking than the Escherichia coli KRX Wildtyp, suggesting an increasing membrane permeability with increasing promotor strength. E. coli with T7 promotor (<bbpart>BBa_K1172502</bbpart>) shows the maximal membrane permeability with 100% enhanced permeability in comparison to Escherichia coli KRX Wildtyp. The weak Anderson promoters seem unsuitable for heterologous expression.
ONPG assay
Another way of characterizing the outer cell membrane can be achieved by ONPG assay. ONPG assay measures the membrane permeability by whole-cell lactase enzyme activity (Zhou et al., 2010).
Electron shuttle-mediated electron transfer (EET) determines the transport efficiency of electron shuttles across the cell membrane. With NPN uptake assay we were able to quantify the transport efficiency of chemical molecules across the cell membrane. ONPG assay (Whole-cell β-lactase enzyme activity assay) can show furthermore the diffusion of ONPG hydrolytic product out of the cell membrane, which quantifies not only uptaking of chemical molecules, but also the hydrolytic product secretion. With ONPG assay we are able to observe diffusion processes in and out of the cell membrane.
Due the fact that heterologous expression of OprF in Escherichia coli has no impact on the expression of β-lactase, we can assume that the β-lactase activity of Escherichia coli KRX Wildtyp and Escherichia coli KRX with OprF expression plasmid is identical. All in all, the ONPG assay provides significantly more information than the NPN uptake assay.
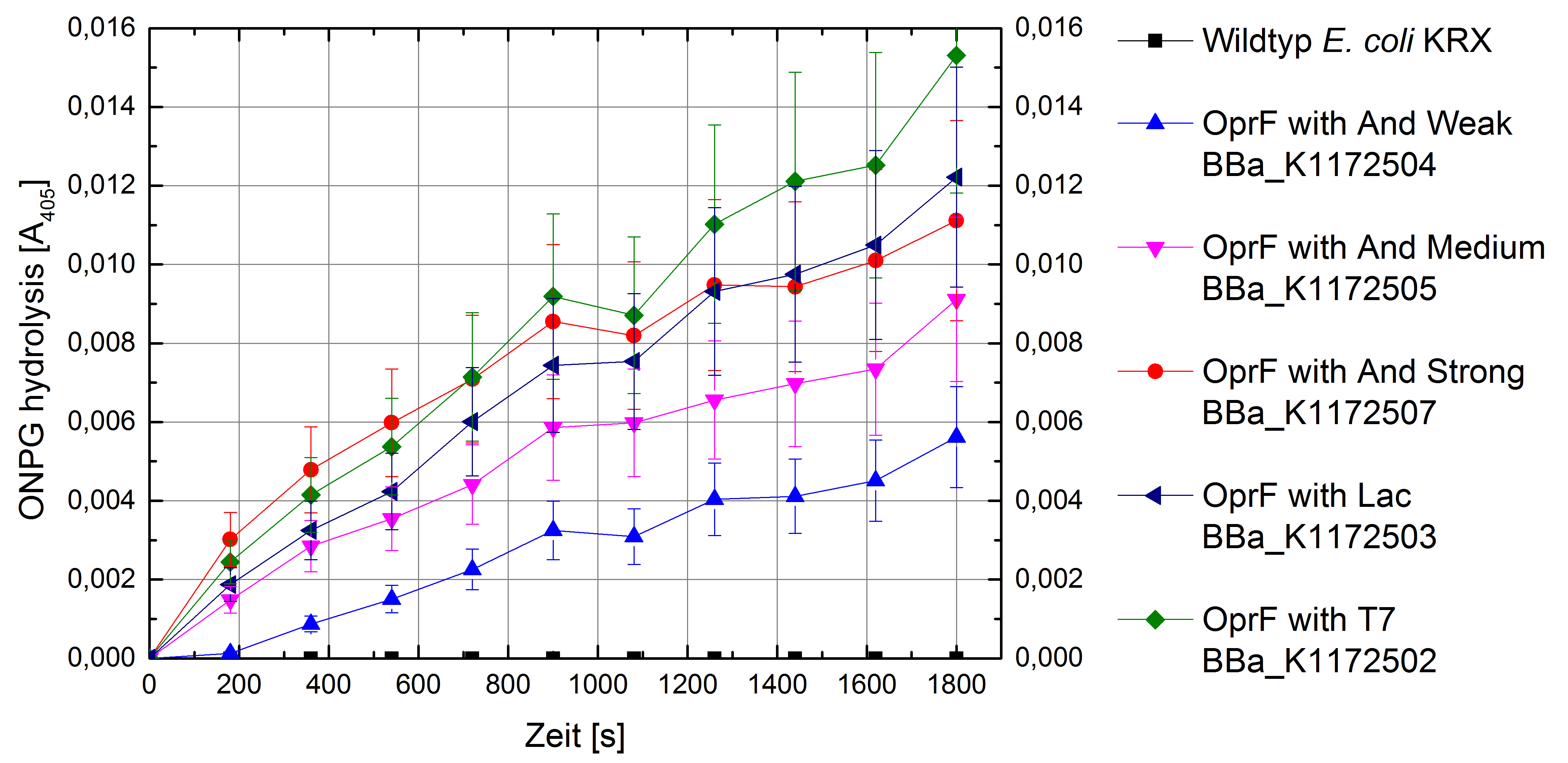
Figure 5: Results of the ONPG-uptake-assay. Comparison of ONPG hydrolysis between Escherichia coli KRX wild type and Escherichia coli KRX with <bbpart>BBa_K1172502</bbpart>, <bbpart>BBa_K1172503</bbpart>, <bbpart>BBa_K1172504</bbpart>, <bbpart>BBa_K1172505</bbpart> and <bbpart>BBa_K1172507</bbpart>. Absorbance at 405 nm wavelength with standard deviation is shown.
The ONPG hydrolysis rate by β-lactase is much higher for Escherichia coli KRX with OprF plasmids in contrast to Escherichia coli KRX Wildtyp. Whereas we couldn’t observe any hydrolysis activity for Wildtyp. Escherichia coli KRX with OprF plasmids shows a much faster ONPG hydrolysis rate with increasing promotor strength and a maximal rate for Escherichia coli KRX with T7 promotor (<bbpart>BBa_K1172502</bbpart>), which is 30 times higher than Wildtyp hydrolysis rate.
In summary we can say that the heterologous expression of OprF from Pseudomonas fluorescens in Escherichia coli significantly improves the membrane permeability. NPN and ONPG assay show correlating results. Escherichia coli outer membrane permeability is enhanced with increasing promotor strength for OprF expression. The Wildtyp shows only a weak uptake rate of chemical molecules (NPN) but no product secretion as quantified with ONPG assay. Therefore, Wildtyp is not suitable for a usage in the MFC. Whereas membrane optimized Escherichia coli with OprF shows great diffusion processes in and out of the cellular membrane, indicating a great optimization of electron shuttle-mediated electron transfer (EET) to the anode and increasing current production.
Atomic Force Microscopy (AFM)
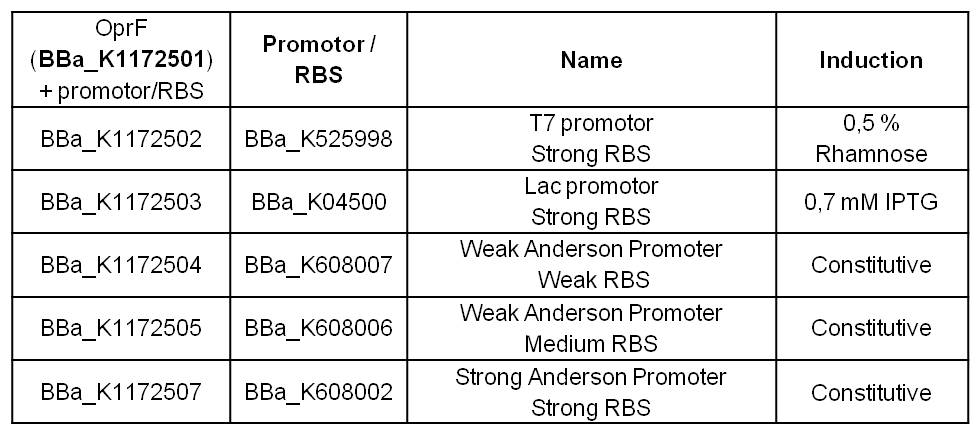
In addition to morphology and physicochemical characterization of the Escherichia coli outer membrane, we wanted to visualize the surface. The technique of choice for this is Atomic Force Microscopy (AFM). After cell preparation we were able to get AFM pictures of E. coli surface with the help of the working group of [http://www.physik.uni-bielefeld.de/biophysik Prof. Dr. Dario Anselmetti], with special help from [http://www.physik.uni-bielefeld.de/biophysik/mitarbeiter/walhorn.html Dr. Volker Walhorn]. Thank you very much for your help!

Figure 6: Microscopy of AFM layer after cell preparation of Escherichia coli KRX Wildtyp.

Figure 7: Microscopy of AFM layer after cell preparation of Escherichia coli with T7 promotor (<bbpart>BBa_K1172502</bbpart>).
Microscopy of AFM glass layer after cell preparation and before AFM measurement shows the different cell properties at the same image enlargement of the microscope and the same number of cells. Escherichia coli KRX with T7 promotor (<bbpart>BBa_K1172502</bbpart>) shows a clustering of cells, whereas wild-type forms a monolayer. The increased hydrophobicity of the cell surface is already visible even with an ordinary light microscope.
AFM was carried out using the [http://www.bruker.com/de/products/surface-analysis/atomic-force-microscopy/multimode-8/overview.html MultiMode® 8 AFM from Bruker]. The measurements were performed on air with ‘Tapping Mode’ and in water with ‘Peak Force Mode’.
According to AFM images, Escherichia coli KRX with OprF and T7 promotor (<bbpart>BBa_K1172502</bbpart>) shows a slightly rougher cell surface morphology in contrast to Escherichia coli KRX Wildtyp. Outer membrane porin OprF from Pseudomonas fluorescens usually forms trimer complex on the membrane, which leads to enhanced roughness of the cell surface. (Yong et al., 2013)
The heterologous expression of porin OprF causes a slightly rougher membrane in which the morphology and thus the viability of Escherichia coli is preserved as far as possible.
Microbial Fuel Cell Measurement
All results described by different evidence indicate an increase in energy production. All these assumptions were confirmed by cultivation of Escherichia coli KRX with OprF (<bbpart>BBa_K1172502</bbpart>) in contrast to Escherichia coli KRX Wildtyp in the Microbial Fuel Cell.
According to our assumptions, the extracellular electron transfer mediated by electron shuttles is improved in the OprF strain resulting in an increased bioelectricity output. (Figure 10 and 11)
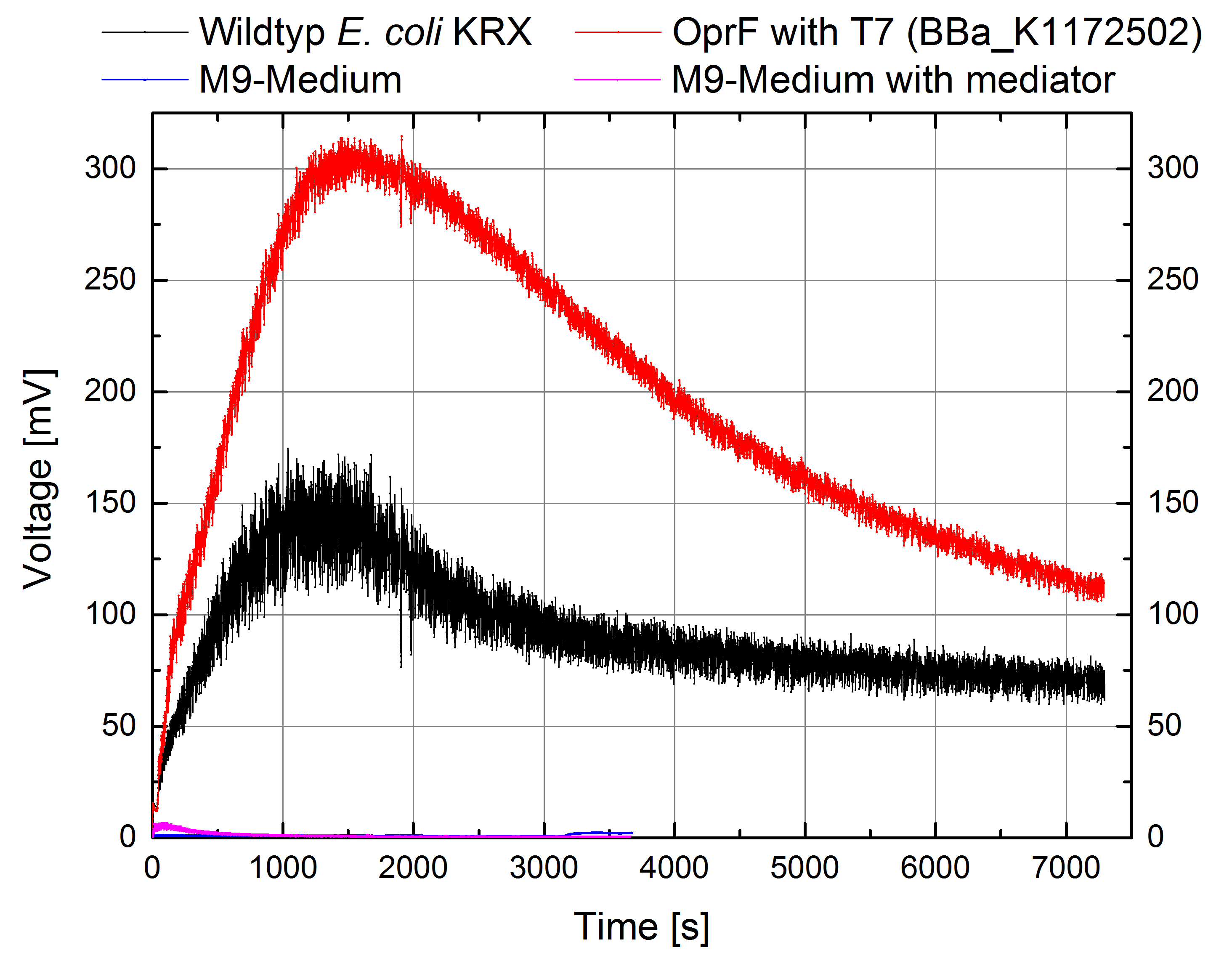
Figure 10: Microbial Fuel Cell results from cultivation of Escherichia coli KRX with OprF (<bbpart>BBa_K1172502</bbpart>) in contrast to Escherichia coli KRX Wildtyp. Voltage curve from Escherichia coli KRX Wildtyp, Escherichia coli KRX with OprF (<bbpart>BBa_K1172502</bbpart>), M9 medium with used mediator New Methylene Blue and M9 medium without mediator is shown over time.
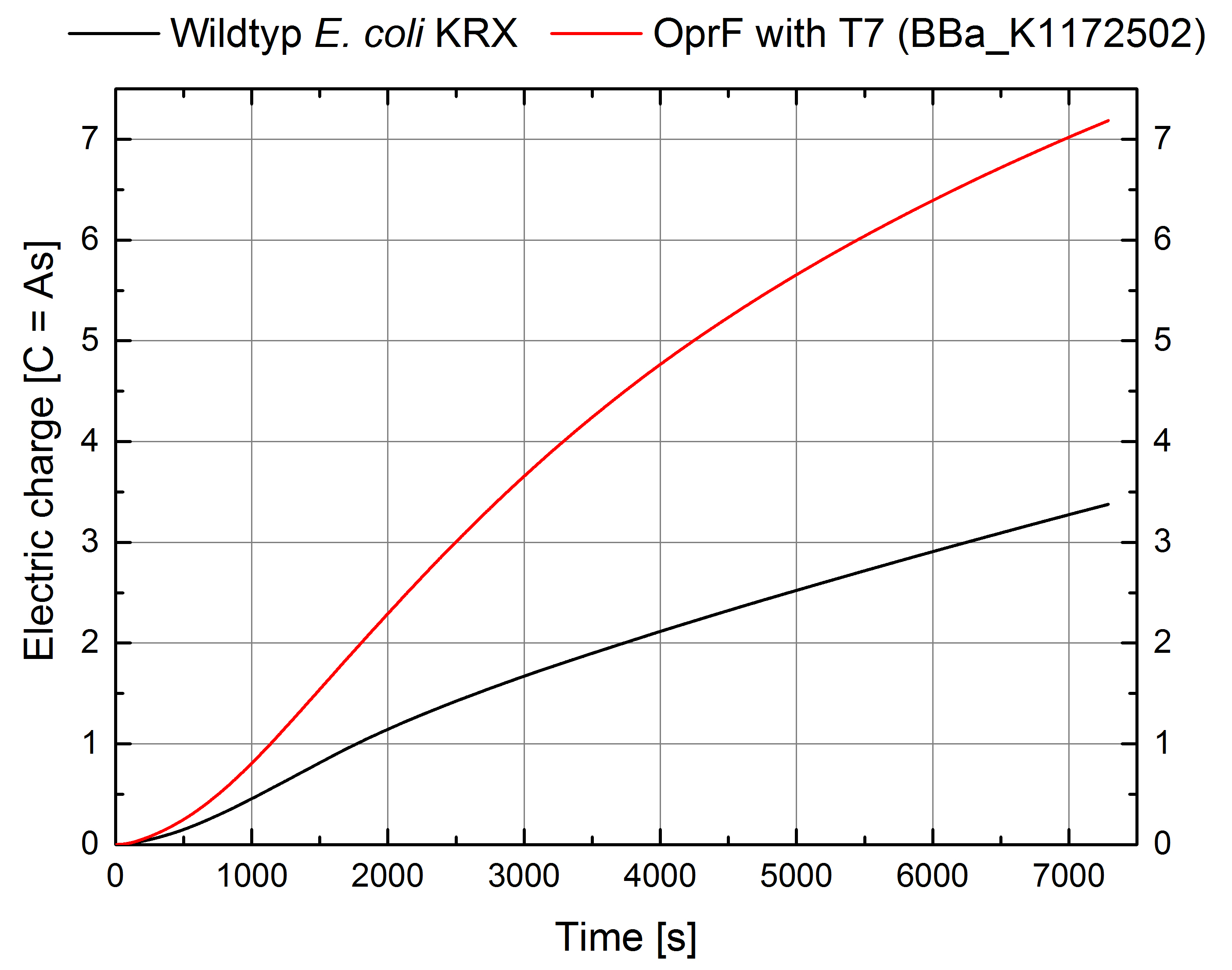
Figure 11: Microbial Fuel Cell results from cultivation of Escherichia coli KRX with OprF (<bbpart>BBa_K1172502</bbpart>) in contrast to Escherichia coli KRX Wildtyp. Electric charge curve from Escherichia coli KRX Wildtyp and Escherichia coli KRX with OprF (<bbpart>BBa_K1172502</bbpart>). M9 medium was used with mediator New Methylene Blue.
Escherichia coli KRX with OprF (<bbpart>BBa_K1172502</bbpart>) shows 108 % higher maximal voltage than Escherichia coli KRX Wildtyp. Over the whole cultivation, voltage was about 100 % improved with maximum at 308 mV. Tests of M9-medium with and without mediator New Methylene Blue shows no voltage, indicating that bioelectricity generation is solely due to the bacteria.
The calculation of the electric charge confirms the described results. Electric charge is equivalent to the number of transported electrons and 111 % enhanced for Escherichia coli KRX with OprF (<bbpart>BBa_K1172502</bbpart>). The maximal electric charge of 7,2 C examines that heterologous expression of outer membrane porin OprF enhances dramatically extracellular electron transfer. High membrane permeability is crucial for efficient mediator transport across the membrane and high bioelectricity generation.
Conclusion
Cell membranes are protective layers which are crucial for physiology of the bacteria. In consideration of the Microbial Fuel Cell, membrane is also a barrier which decreases mediator and thus electron exchange between bacteria and anode. With heterologous expression of the outer membrane porin OprF from Pseudomonas fluorescens in Escherichia coli we are able to enhance membrane permeability. With this optimized cell membrane surface we generate an efficient electron shuttle-mediated EET with decreased limitation of EET by membrane barrier. In contrast to a perforation of the membrane with chemicals, cell viability is maintained with OprF expression (Liu et al., 2012). Besides improvement of EET, enhanced hydrophobicity shows optimized cell adhesion to the anode for biofilm formation and direct electron transfer.
The heterologous expression of OprF is a great genetic strategy to optimize electron shuttle-mediated electron transfer as well as electricity generation in Microbial Fuel Cells. The most suitable and efficient OprF device for Escherichia coli is a combination with Rhamnose inducible T7 promotor (<bbpart>BBa_K1172502</bbpart>).
References
Hancock REW, Brinkman FSL (2002) Function of Pseudomonas porins in uptake and efflux. [http://www.annualreviews.org/doi/abs/10.1146/annurev.micro.56.012302.160310| Annu Rev Microbiol (56):17–38].
Helander IM, Mattila-Sandholm T (2000) Permeability barrier of the gramnegative bacterial outer membrane with special reference to nisin. [http://www.sciencedirect.com/science/article/pii/S016816050000307X|Int J Food Microbiol 60: 153–161].
Liu J, Qiao Y, Lu ZS, Song H, Li CM (2012) Enhance electron transfer and performance of microbial fuel cells by perforating the cell membrane. [http://www.sciencedirect.com/science/article/pii/S1388248111004723|Electrochem Commun 15: 50–53].
Logan BE (2009) Exoelectrogenic bacteria that power microbial fuel cells. [http://www.nature.com/nrmicro/journal/v7/n5/full/nrmicro2113.html| Nat Rev Microbiol (7): 375–381].
Loh B, Grant C, Hancock REW (1984) Use of the fluorescent-probe 1-Nphenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer-membrane of Pseudomonas aeruginosa. [http://aac.asm.org/content/26/4/546.short|Antimicrob Agents Chemother 26: 546–551].
Yong YC, Yu YY, Yang Y, Liu J, Wang JY, Song H (2013) Enhancement of Extracellular Electron Transfer and Bioelectricity Output by Synthetic Porin. [http://onlinelibrary.wiley.com/doi/10.1002/bit.24732/full|Biotechnology and Bioengineering 110 (2): 408-416].
Zhou ZX, Wei DF, Guan Y, Zheng AN, Zhong JJ (2010) Damage of Escherichia coli membrane by bactericidal agent polyhexamethylene guanidine hydrochloride: Micrographic evidences. [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2009.04482.x/full|J Appl Microbiol 108: 898–907].
 "
"