Team:Warsaw/Acrylamide detection
From 2013.igem.org
| Line 13: | Line 13: | ||
[[File:Hemo.png|600px|thumb|left|A simplified overview of the hemoglobin-based sensor.]] | [[File:Hemo.png|600px|thumb|left|A simplified overview of the hemoglobin-based sensor.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
Revision as of 22:14, 4 October 2013
Acrylamide detection
Contents |
Hemoglobin-based sensor
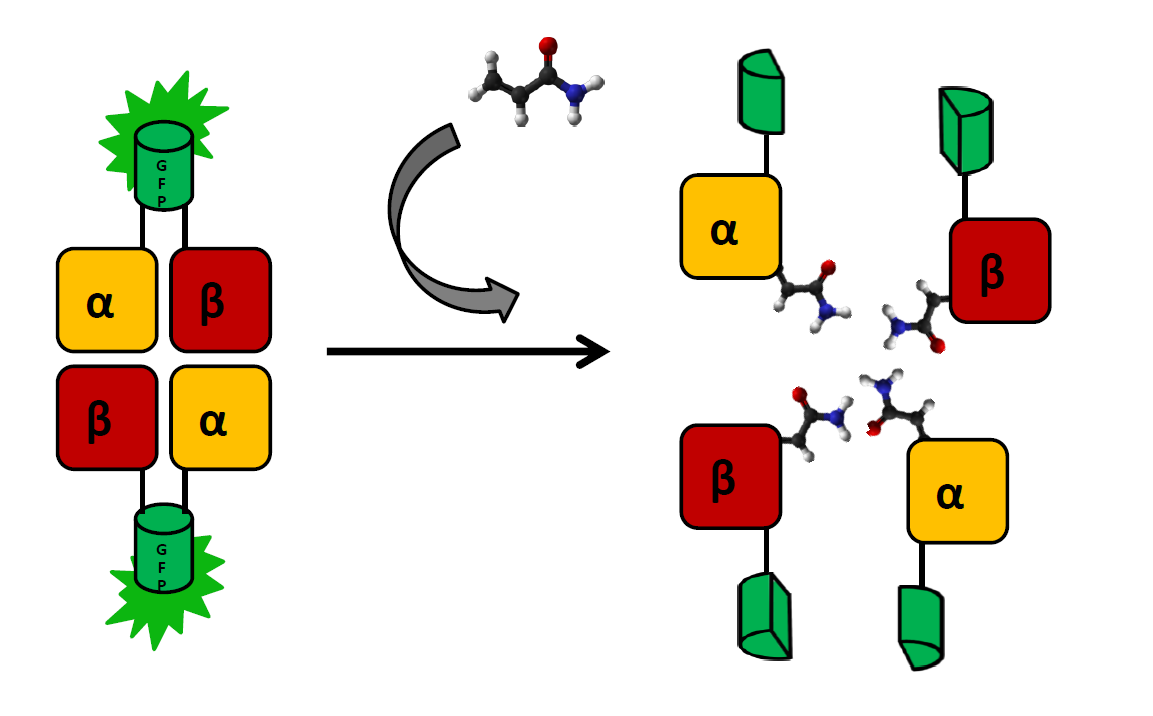
The main indicator of exposure to acrylamide are adducts formed at the N-terminal valine of both hemoglobin chains. These can (and have been) detected by coating a glass electrode with human hemoglobin an measuring changes in intensity to voltage ratio (Krajewska, A. et al.). We wish to create a cheaper and simpler variant of this detection system by applying E. coli strain that produces functional human hemoglobin (a modification of BactoBlood system created by Berkley 2007) with both α and β chains fused with split fluorophore fragments derived from sfGFP (superfolder GFP) protein (Zhou, J. et al.) at the C-terminus. Constructs expressing each fusion protein would be transformed into two separate E. coli cultures, which would then be exposed to a solution containing acrylamide and lysed by inducing one of two kill-switches designed this year (see: Safety).
As the presence of acrylamide adducts at the N-terminus of hemoglobin chains results in a decrease of mutual affinity, it is also expected to lead to a significantly weaker fluorescence in relation to controls. There will be two controls negative:
- detection system in a solution without acrylamide
- bJun/bFos proteins (see: BiFC toolbox) fused with the same split fluorophore fragments and exposed to the same acrylamide solution. Interaction between those proteins should not be affected by acrylamide
Subunits will be expressed separately, as the formation of functional fluorophore is generally irreversible. sfGFP was chosen for this system, as superfolder mutations result in better resistance to unspecific quenching:
roGFP-based sensor
In parralel we planned to develop a separate method of detection, based on a redox state reporter - roGFP protein. A fusion of roGFP with human glutaredoxin is a highly specifc reporter of redox potential of cellular glutathione pool (Gutscher, M. et al.). Considering the fact that conjugation with glutathione (by glutathione S-tranferase) is a key pathway of detoxication following acrylamide exposure, its effect on the redox state of gluthatione pool should be easily observable (Cui, S. et al.). roGFP is a derivative of GFP with two additional cysteines introduced via site-directed mutagenesis. Depending on the redox state, they can form a disulfide bond. This leads to a shift in excitation peak. This effect is more reliable than mere decrease in fluorescence and requires only one simple negative control. As the measurements will be conducted in E. coli cells, we designed two fusion proteins roGFP with human glutaredoxin and with E. coli glutaredoxin. Unfortunately, due to delays in gene synthesis, we weren't able to test either of the detection systems in time before wiki freeze.
| Tertiary structure of roGFP with two possible redox states of the cysteines introduced by site-directed mutagenesis |
Bibliography:
- Krajewska, A., Radecki, J. & Radecka, H. A Voltammetric Biosensor Based on Glassy Carbon Electrodes Modified with Single-Walled Carbon Nanotubes/Hemoglobin for Detection of Acrylamide in Water Extracts from Potato Crisps. Sensors 8, 5832–5844 (2008).
- Berkeley_UC @ 2007.igem.org. at [1]
- Zhou, J., Lin, J. & Zhou, C. An improved bimolecular fluorescence complementation tool based on superfolder green fluorescent protein. Acta biochimica et … 43, 239–244 (2011).
- Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology 24, 79–88 (2006).
- Hanson, G. T. et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. The Journal of biological chemistry 279, 13044–53 (2004).
- Gutscher, M. et al. Real-time imaging of the intracellular glutathione redox potential. Nature methods 5, 553–9 (2008).
- Cui, S., Kim, S., Jo, S. & Lee, Y. A study of in vitro scavenging reactions of acrylamide with glutathione using electrospray ionization tandem mass spectrometry. BULLETIN-KOREAN … (2005).at [http://newjournal.kcsnet.or.kr/main/j_search/j_download.htm?code=B050816]
 "
"