Team:ATOMS-Turkiye/Project/Module2/Experiment
From 2013.igem.org
(→Challenges:) |
(→Design and assembly) |
||
| Line 6: | Line 6: | ||
=Design and assembly= | =Design and assembly= | ||
| - | We aim to ensure that | + | ===Apoptin and E4of4=== |
| - | In | + | We aim to ensure that the specifications drawn are considered in the design. |
| - | + | In recent studies, Apoptin has been described to induce apoptosis in various human cancer cell lines. To develop apoptin as an anti-cancer drug, efficient transduction tools have been used to deliver this protein into the tumor cells. Cell-penetrating peptides, such as the trans-acting activator of transcription (TAT) protein transduction domain (PTD) and MPG peptide have therefore been used as vectors for protein delivery. Taking on from this, we decided to fuse apoptin with TAT peptide as it has the ability to penetrate cells without causing any membrane damage. On the one hand, we also fused Hemoglutinin A with TAT in order to decrease possibility of getting trapped in microcytosis.In addition to this, we conceived an alternative technology which offers fusing another cell penetrating peptide with apoptin, thus we constructed MPG peptide with apoptin. We applied this idea again, with another protein named as E4of4. E4of4 is a tumor specific molecule and is known to induce apoptosis. We demonstrated e4of4 fused to TAT peptide as an alternative to apoptin which also aims at tumor cells. Therefore, we added MPG-Apoptin and TAT-E4of4 into literature. | |
| - | + | We ordered apoptin fused to TAT, TAT HA and MPG sequences respectively as a composite part in order to minimalize cost and time. We cloned our parts into pSB1C3 backbone. Once cloned, first our proteins were expressed in BL21 e. coli strain under constutive promoter.In addition to this,our constructs were harboring a 6xHis tag.The 6×His were fused with constructs at the N-terminus to enhance purification. | |
| - | + | After cultivatization, we examined the results of our genes using gel electrophoresis. The electrophoresis proved that the transformation of new genes were successful.The results of our gel electrophoresis are below: | |
| - | After cultivatization, we | + | |
| - | + | ||
Experiments and results: | Experiments and results: | ||
| - | To purify the recombinant proteins, cells were spun down from 50 mL of culture supernatant and resuspended in phosphate buffer saline.PBS contained PMSF in order to prevent proteis from proteases. The mixture was then sonicated on ice eight times for 30 second with a 52% pulsed activity cycle (MISONIX Sonicator W 300). Next, the lysate was centrifuged for 30 min at 10,000 rpm to remove the cell debris. The resulting cell supernatant was loaded onto HIS select affinity column (Sİgma Aldrich)) for protein purification using the standard procedure | + | To purify the recombinant proteins, cells were spun down from 50 mL of culture supernatant and resuspended in phosphate buffer saline.PBS contained PMSF in order to prevent proteis from proteases. The mixture was then sonicated on ice eight times for 30 second with a 52% pulsed activity cycle (MISONIX Sonicator W 300). Next, the lysate was centrifuged for 30 min at 10,000 rpm to remove the cell debris. The resulting cell supernatant was loaded onto HIS select affinity column (Sİgma Aldrich)) for protein purification using the standard procedure. The total protein concentration of each collected fraction from the column was determined using Bradford protein assay with bovine serum albumin acting as the reference protein. The purity of the protein from each fraction was analyzed by 12.5% SDS-PAGE and then the resulting gels were Western blotted using monoclonal anti-HIS antibody. |
| - | monoclonal anti-HIS antibody. | + | |
| - | Using western blot analysis, it was observed that; our proteins expressed and purified effectively. | + | Using western blot analysis, it was observed that; our proteins were expressed and purified effectively. |
| - | Firstly, | + | Firstly, we offered to use Immunofluorescence staining method to understand the localisation of TAT fusing proteins.Therefore,we fused TAT-RFP construct with 6xHis tag in order to prove TAT's activity. Immunofluorescence is a technique used for light microscopy with a fluorescence microscope and is used primarily on microbiological samples. This technique uses the specificity of antibodies to their antigen in order to target fluorescent dyes to specific biomolecule targets within a cell, therefore allowing the visualisation of the distributed target molecules in the sample.When Flourochrome labeled anti Histag antibody bonded to 6His-tag located in front of our construct, we were able to visualize TAT-RFP in the intracellular area. We also stained DNA and nuclei using Hoechst dye (Thermo Scientific Pierce Hoechst 33342 Fluorescent Stain ) to ensure that our TAT peptide was able to penetrate the cells. |
| - | + | We investigated whether his-TAT-Apoptin, TAT-HA-Apoptin, TAT-E4of4 and MPG-Apoptin protein expressed by E. coli is able to induce apoptotic activity when introduced into tumor cells. Purified protein was used to examine the protein's apoptotic activity to treat human colon cancer HT29, breast cancer MCF7 and head-neck cancer cell lines. We repeated the experiments by changing our variables such as dosage, cell line, changing purified methods and analysed cell viability index under changing factors.We used xCelligence for analysis. The xCELLigence System, also known as RT-CES system, which contains a series of Real Time Cell Analyzer (RTCA), is a labeling free cell based assay system integrating microelectronics and cell biology, suitable for uninterrupted monitoring of biolgical processes of living cells. | |
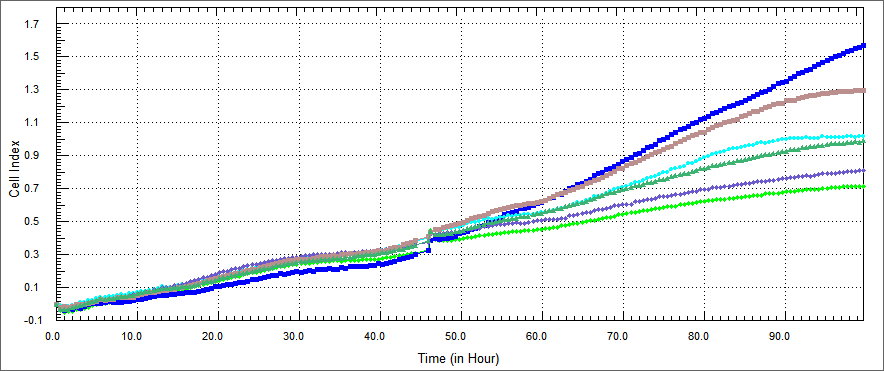
| - | We conducted experiments on in vitro cultured HT29 cells which were treated with TAT-Apoptin, TAT-E4of4, TAT-HA-Apoptin and MPG-Apoptin. The results of the X-Cellingence revealed that our constructs | + | We conducted experiments on in vitro cultured HT29 cells which were treated with TAT-Apoptin, TAT-E4of4, TAT-HA-Apoptin and MPG-Apoptin. The results of the X-Cellingence revealed that our constructs were able to significantly inhibit the growth of the HT29 cells. We used DMEM and elution buffer containing well as a control. |
[[File:Oncoli-exp1-ht29 1.png|800px]] | [[File:Oncoli-exp1-ht29 1.png|800px]] | ||
| - | + | The growth rate of HT 29 cells were performed by xCELLigence after %5 concentration of the purified E. coli-expressed recombinant proteins were co-cultured with HT-29 cell for 54 hours. Non-apoptosis controls, only DMEM and elution buffer treated cells were assayed. These respectively represent HL-60 cells only (blue), HT-29 cells co-cultured with protein elution buffer (brown) and HT-29 cells co-cultured with TAT-Apoptin (light green),TAT-HA-Apoptin (dark green), MPG-Apoptin (light blue) and TAT-E4of4(purple). | |
| - | + | The results were observed in a certain range of concentration and time, our proteins inhibited the proliferation of the HT 29 cells time dependently. Its inhibitory effect was %5 concentration and 54 hours. | |
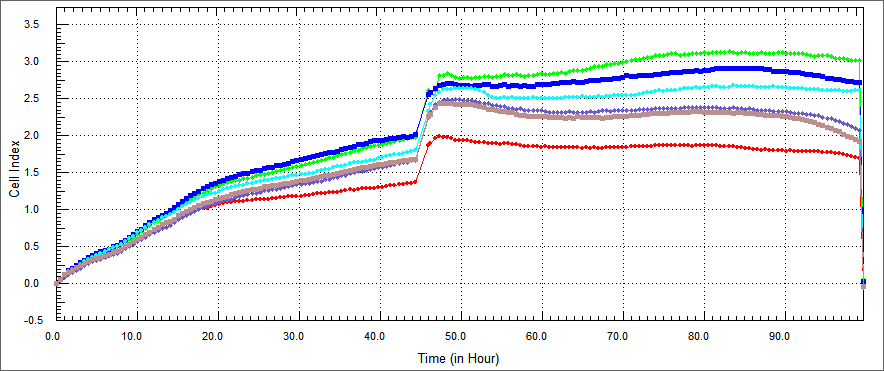
| - | Using a similar approach, we tested the apoptosis-inducing ability | + | Using a similar approach, we tested the apoptosis-inducing ability of our constructs in HEK 293 human kidney cell lines.HEK 293 were co-cultured with TAT-Apoptin, TAT-E4of4, TAT-HA-Apoptin and MPG-Apoptin.In a desired concetration; we expect that; growth curve of treated with our construct HEK 293 cells should be similary growth curve of not treated HEK 293. |
[[File:Oncoli-exp1-hek1.png|800px]] | [[File:Oncoli-exp1-hek1.png|800px]] | ||
| Line 58: | Line 55: | ||
| - | + | We also thought about alternative ways to demonstrate apoptosis and decided to do XTT assay. XTT assay is a colorimetric assay for the nonradioactive quantification of cellular proliferation, viability, and cytotoxicity.We measured spectrophometric analysis by ELIZA. The improved XTT method was used to study the effect of our constructs on the growth of colon, and head-neck cancer cells. The cell concentration was adjusted to 5x104 cells/mland the cells were added to a 96-well plate, at 90 µl/well. There was %10 concentrations of each constructs and each protein was set in three parallel wells with 10 µl/well. The cells were then cultured at 37˚C in a 5% (volume fraction) CO 2incubator and incubated for 48 h. XTT (5 mg/ml) was then added at 50 µl/well and the cells were cultured for 4 h. Following a period of 24, 48 or 72 h, cell viability was determined by a microplate reader at 5 nm single wavelength optical density (OD). | |
Latest revision as of 15:46, 19 May 2014
 "
"